Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
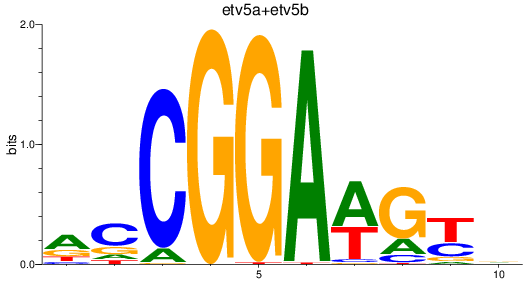
Results for etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
Z-value: 5.08
Transcription factors associated with etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv5b
|
ENSDARG00000044511 | ETS variant transcription factor 5b |
|
etv5a
|
ENSDARG00000069763 | ETS variant transcription factor 5a |
|
etv5a
|
ENSDARG00000113729 | ETS variant transcription factor 5a |
|
etv5b
|
ENSDARG00000113744 | ETS variant transcription factor 5b |
|
elk1
|
ENSDARG00000078066 | ETS transcription factor ELK1 |
|
elk4
|
ENSDARG00000077092 | ETS transcription factor ELK4 |
|
etv1
|
ENSDARG00000101959 | ETS variant transcription factor 1 |
|
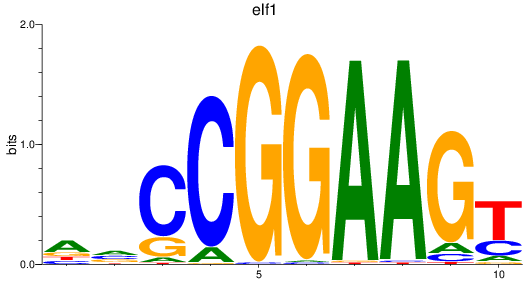
elf1
|
ENSDARG00000020759 | E74-like ETS transcription factor 1 |
|
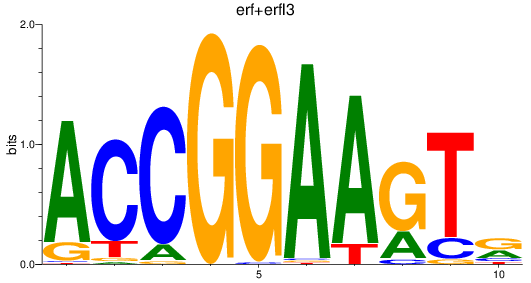
erfl3
|
ENSDARG00000062801 | Ets2 repressor factor like 3 |
|
erf
|
ENSDARG00000063417 | Ets2 repressor factor |
|
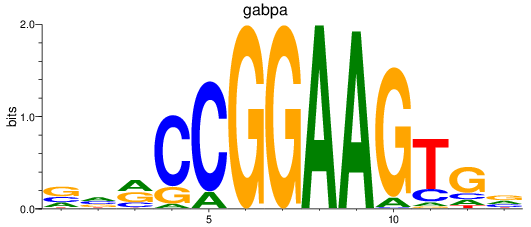
gabpa
|
ENSDARG00000069289 | GA binding protein transcription factor subunit alpha |
|
gabpa
|
ENSDARG00000110923 | GA binding protein transcription factor subunit alpha |
|
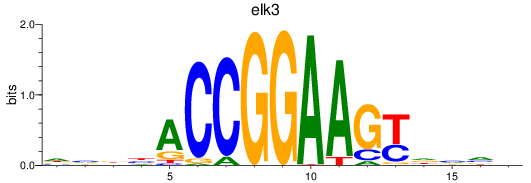
elk3
|
ENSDARG00000018688 | ETS transcription factor ELK3 |
|
elk3
|
ENSDARG00000110853 | ETS transcription factor ELK3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| etv1 | dr11_v1_chr15_-_34214440_34214440 | -0.99 | 2.0e-07 | Click! |
| erf | dr11_v1_chr19_-_6193067_6193141 | 0.95 | 7.7e-05 | Click! |
| etv5a | dr11_v1_chr9_+_22634073_22634073 | 0.91 | 7.5e-04 | Click! |
| elf1 | dr11_v1_chr14_-_40797117_40797117 | 0.88 | 1.5e-03 | Click! |
| gabpa | dr11_v1_chr9_+_35015747_35015747 | 0.87 | 2.6e-03 | Click! |
| elk3 | dr11_v1_chr4_+_7698862_7698862 | -0.85 | 3.4e-03 | Click! |
| etv5b | dr11_v1_chr6_-_14040136_14040161 | 0.79 | 1.2e-02 | Click! |
| erfl3 | dr11_v1_chr16_+_11029762_11029762 | -0.75 | 2.0e-02 | Click! |
| elk1 | dr11_v1_chr8_+_8712446_8712446 | 0.68 | 4.5e-02 | Click! |
| elk4 | dr11_v1_chr11_-_38492269_38492269 | 0.64 | 6.5e-02 | Click! |
Activity profile of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3 motif
Sorted Z-values of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_8365398 | 8.00 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr9_+_8408054 | 7.07 |
ENSDART00000144373
|
zgc:153499
|
zgc:153499 |
| chr1_-_6085750 | 6.27 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr12_-_10508952 | 5.91 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr8_-_33154677 | 5.35 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr8_-_36554675 | 5.25 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr7_+_55518519 | 5.21 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr14_+_22457230 | 5.12 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr1_+_59314675 | 5.04 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr22_+_35275468 | 5.03 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr9_+_8407778 | 5.02 |
ENSDART00000102754
ENSDART00000178144 |
zgc:153499
|
zgc:153499 |
| chr2_-_57900430 | 5.02 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr4_+_279669 | 4.99 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr13_-_33700461 | 4.93 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr5_-_26795438 | 4.91 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr5_+_3927989 | 4.91 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr15_+_34933552 | 4.88 |
ENSDART00000155368
|
zgc:66024
|
zgc:66024 |
| chr2_+_30182431 | 4.83 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr5_-_13206878 | 4.80 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr7_+_74141297 | 4.65 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr9_-_10805231 | 4.64 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr3_+_32416948 | 4.60 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr22_+_35275206 | 4.51 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr2_-_31711547 | 4.42 |
ENSDART00000175525
ENSDART00000189763 ENSDART00000113498 |
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr21_-_30994577 | 4.34 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr8_+_36554816 | 4.34 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr3_-_15131438 | 4.31 |
ENSDART00000131720
|
xpo6
|
exportin 6 |
| chr14_-_14687004 | 4.28 |
ENSDART00000169970
|
gcna
|
germ cell nuclear acidic peptidase |
| chr21_-_36453594 | 4.27 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr22_-_11829436 | 4.17 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr20_+_13141408 | 4.15 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr25_-_19574146 | 4.13 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr11_-_36230146 | 4.12 |
ENSDART00000135888
ENSDART00000189782 |
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr25_+_186583 | 4.12 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr14_+_16083818 | 4.08 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr13_-_24396003 | 4.08 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr12_+_47794089 | 4.06 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr17_-_868004 | 4.06 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr10_+_17747880 | 4.05 |
ENSDART00000135044
|
pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr9_-_7287375 | 4.02 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr8_-_20862443 | 4.01 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr22_-_10752471 | 3.89 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr12_-_35582683 | 3.87 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr13_-_35760969 | 3.86 |
ENSDART00000127476
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr15_-_5892650 | 3.85 |
ENSDART00000154812
|
brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr14_-_26465729 | 3.84 |
ENSDART00000143454
|
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr15_-_43625549 | 3.83 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr13_-_24396199 | 3.82 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr5_+_28497956 | 3.81 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr20_-_46866934 | 3.75 |
ENSDART00000158178
|
si:ch73-21k16.4
|
si:ch73-21k16.4 |
| chr21_+_26028947 | 3.73 |
ENSDART00000028007
|
supt6h
|
SPT6 homolog, histone chaperone |
| chr9_-_7287128 | 3.70 |
ENSDART00000176281
ENSDART00000065803 |
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr25_-_21782435 | 3.69 |
ENSDART00000089616
|
bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr1_-_8553165 | 3.68 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr8_+_23726708 | 3.67 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr2_-_57918314 | 3.65 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr16_+_25259313 | 3.62 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr12_-_35582521 | 3.60 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr11_-_34480822 | 3.59 |
ENSDART00000129029
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr9_-_12269847 | 3.58 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr19_+_19241372 | 3.56 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr3_+_59880317 | 3.55 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr2_+_38055529 | 3.55 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr12_+_10443785 | 3.50 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr13_-_35761266 | 3.49 |
ENSDART00000190217
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr6_-_54433995 | 3.48 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr1_-_34685329 | 3.48 |
ENSDART00000125944
ENSDART00000008277 |
pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr1_-_55044256 | 3.47 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr22_-_38274188 | 3.47 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr16_+_54674556 | 3.47 |
ENSDART00000167040
|
pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr17_+_50261603 | 3.46 |
ENSDART00000154503
ENSDART00000154467 |
syncripl
|
synaptotagmin binding, cytoplasmic RNA interacting protein, like |
| chr7_+_13684012 | 3.46 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr25_+_3549841 | 3.44 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr8_+_8643901 | 3.44 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr23_-_37113396 | 3.44 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr2_-_4070850 | 3.43 |
ENSDART00000159990
|
yme1l1b
|
YME1-like 1b |
| chr15_-_34933560 | 3.42 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr15_-_14642186 | 3.42 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr13_+_9559461 | 3.39 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr14_-_45558490 | 3.38 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
| chr13_+_15933168 | 3.37 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr9_+_426392 | 3.34 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr21_-_36453417 | 3.33 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr10_-_36547128 | 3.28 |
ENSDART00000108484
ENSDART00000086861 |
nipblb
|
nipped-B homolog b (Drosophila) |
| chr4_-_858434 | 3.28 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr21_+_19925910 | 3.26 |
ENSDART00000111694
ENSDART00000132653 |
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr8_+_23726244 | 3.26 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr11_-_45385803 | 3.26 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr6_+_4387150 | 3.18 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr10_-_4961923 | 3.17 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr25_+_35889102 | 3.13 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr7_+_38744924 | 3.12 |
ENSDART00000052329
|
znf408
|
zinc finger protein 408 |
| chr3_-_36602379 | 3.09 |
ENSDART00000161501
ENSDART00000162396 |
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr14_-_10617923 | 3.09 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr11_+_18183220 | 3.09 |
ENSDART00000113468
|
LO018315.10
|
|
| chr14_+_11289851 | 3.07 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr6_+_41808673 | 3.05 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr14_-_899170 | 3.03 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr9_-_50001606 | 3.02 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_+_25171832 | 3.01 |
ENSDART00000156416
|
wu:fe05a04
|
wu:fe05a04 |
| chr2_+_34112100 | 3.01 |
ENSDART00000056666
ENSDART00000146624 |
klhl20
|
kelch-like family member 20 |
| chr21_-_41369539 | 3.00 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr1_+_494297 | 2.99 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr18_-_20458412 | 2.98 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr5_-_23485161 | 2.98 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr4_+_5318901 | 2.97 |
ENSDART00000067378
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr14_+_32918484 | 2.96 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr16_+_38337783 | 2.96 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr25_-_36263115 | 2.94 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr7_-_69298271 | 2.93 |
ENSDART00000122935
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr5_-_4297459 | 2.92 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr15_-_41677689 | 2.91 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr25_-_12412704 | 2.91 |
ENSDART00000168275
|
det1
|
DET1, COP1 ubiquitin ligase partner |
| chr11_-_16115804 | 2.91 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr14_-_46198373 | 2.90 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr3_-_48612078 | 2.87 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr19_-_29853402 | 2.87 |
ENSDART00000024292
ENSDART00000188508 |
txlna
|
taxilin alpha |
| chr7_+_24573721 | 2.85 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr22_-_11833317 | 2.85 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr16_-_54498109 | 2.85 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr20_-_45772306 | 2.84 |
ENSDART00000062092
|
trmt6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr20_-_48898371 | 2.84 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr18_+_44849809 | 2.84 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_-_69482891 | 2.83 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr2_-_48298985 | 2.81 |
ENSDART00000057957
|
itm2cb
|
integral membrane protein 2Cb |
| chr3_-_30186296 | 2.80 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr13_-_24745288 | 2.80 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr18_+_26429428 | 2.79 |
ENSDART00000142686
|
blm
|
Bloom syndrome, RecQ helicase-like |
| chr22_+_18316144 | 2.79 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr7_+_53254234 | 2.78 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr3_-_5433392 | 2.78 |
ENSDART00000159308
|
trim35-7
|
tripartite motif containing 35-7 |
| chr13_+_49727333 | 2.78 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr4_-_5371639 | 2.77 |
ENSDART00000150697
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr25_+_4855549 | 2.76 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr8_+_15277874 | 2.72 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr20_-_33174899 | 2.71 |
ENSDART00000047834
|
nbas
|
neuroblastoma amplified sequence |
| chr1_-_34685007 | 2.70 |
ENSDART00000157471
|
klf5a
|
Kruppel-like factor 5a |
| chr25_-_24247584 | 2.70 |
ENSDART00000046349
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr12_-_9468618 | 2.69 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr22_-_26289549 | 2.68 |
ENSDART00000043774
|
sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr14_-_51014292 | 2.68 |
ENSDART00000029797
|
faf2
|
Fas associated factor family member 2 |
| chr11_+_24348425 | 2.64 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr4_-_72287764 | 2.63 |
ENSDART00000125452
ENSDART00000189437 |
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr22_+_8753365 | 2.63 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr20_+_54312970 | 2.63 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr18_-_14274803 | 2.62 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr24_-_36196077 | 2.62 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr9_-_15789526 | 2.61 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr15_+_518092 | 2.59 |
ENSDART00000154594
|
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr3_-_36602069 | 2.58 |
ENSDART00000165414
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr18_-_20458840 | 2.58 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr25_-_18948816 | 2.57 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr11_+_25693395 | 2.57 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr16_+_26680508 | 2.53 |
ENSDART00000142215
ENSDART00000159064 |
virma
|
vir like m6A methyltransferase associated |
| chr12_-_3840664 | 2.53 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr25_+_3549401 | 2.52 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr20_-_48898560 | 2.51 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr18_-_21910991 | 2.51 |
ENSDART00000089787
ENSDART00000169220 ENSDART00000132381 ENSDART00000191764 |
edc4
|
enhancer of mRNA decapping 4 |
| chr1_+_51615672 | 2.51 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr25_+_33202294 | 2.51 |
ENSDART00000131098
|
TPM1 (1 of many)
|
zgc:171719 |
| chr21_-_7781555 | 2.50 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr20_-_48470599 | 2.48 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr23_-_20177527 | 2.47 |
ENSDART00000147778
|
usp19
|
ubiquitin specific peptidase 19 |
| chr11_+_3211826 | 2.47 |
ENSDART00000191419
ENSDART00000188786 ENSDART00000192914 ENSDART00000112417 |
timeless
|
timeless circadian clock |
| chr9_-_49964810 | 2.45 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr20_+_54304800 | 2.45 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr22_+_8753092 | 2.44 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr8_+_7778770 | 2.44 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr24_+_31459715 | 2.43 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr20_+_54299419 | 2.43 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr12_-_2869565 | 2.43 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr18_-_3552414 | 2.43 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr13_-_29980215 | 2.42 |
ENSDART00000042049
|
hif1an
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr6_+_27275716 | 2.42 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr13_-_18691041 | 2.42 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr17_+_39790388 | 2.40 |
ENSDART00000149488
|
ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr19_-_34742440 | 2.40 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr17_+_8754020 | 2.40 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr7_+_22796723 | 2.39 |
ENSDART00000077395
ENSDART00000146368 |
rbm4.2
|
RNA binding motif protein 4.2 |
| chr2_+_5406236 | 2.38 |
ENSDART00000154167
|
sft2d3
|
SFT2 domain containing 3 |
| chr24_-_41195068 | 2.38 |
ENSDART00000121592
|
acvr2ba
|
activin A receptor type 2Ba |
| chr20_+_54309148 | 2.38 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr21_+_3941758 | 2.37 |
ENSDART00000181345
|
setx
|
senataxin |
| chr13_-_12667220 | 2.35 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr21_+_39197628 | 2.35 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr25_+_34246625 | 2.35 |
ENSDART00000082578
|
bnip2
|
BCL2 interacting protein 2 |
| chr5_-_33935396 | 2.35 |
ENSDART00000133578
|
si:dkeyp-72a4.1
|
si:dkeyp-72a4.1 |
| chr14_-_41272034 | 2.35 |
ENSDART00000191709
ENSDART00000074438 |
cenpi
|
centromere protein I |
| chr21_+_20396858 | 2.32 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr20_-_2641233 | 2.32 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr7_+_32693890 | 2.32 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr5_-_25733745 | 2.31 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr21_+_45717930 | 2.31 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr21_-_41369370 | 2.31 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr15_-_11683529 | 2.30 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr16_+_33931032 | 2.29 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr16_-_17345377 | 2.29 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr1_+_40034061 | 2.28 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr19_+_7424347 | 2.27 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr20_-_9123052 | 2.26 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 1.7 | 5.1 | GO:0030237 | female sex determination(GO:0030237) |
| 1.6 | 4.8 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.4 | 8.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 1.4 | 5.5 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 1.3 | 9.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 1.3 | 5.1 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.2 | 3.5 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.1 | 4.5 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.1 | 4.2 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.0 | 10.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.9 | 2.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 5.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.8 | 2.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.8 | 5.3 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.7 | 6.7 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.7 | 8.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.7 | 5.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.7 | 4.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.7 | 2.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.7 | 9.0 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.7 | 2.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.7 | 2.7 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 10.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.7 | 2.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.6 | 2.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.6 | 3.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.6 | 4.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.6 | 3.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.6 | 6.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.6 | 3.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.6 | 1.7 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.6 | 1.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.6 | 1.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 2.2 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.5 | 2.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.5 | 2.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.5 | 1.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 2.6 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 1.6 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.5 | 1.5 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 4.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.5 | 8.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.5 | 1.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 | 1.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.5 | 1.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.5 | 1.5 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.5 | 2.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.5 | 2.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 2.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 1.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 0.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 3.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.4 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 5.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 1.8 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.4 | 9.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.4 | 5.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 3.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 2.6 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 6.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 1.2 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.4 | 3.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.4 | 1.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.4 | 1.6 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.4 | 1.6 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.4 | 1.6 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.4 | 3.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.4 | 0.8 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.4 | 3.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 4.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.4 | 1.8 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.4 | 1.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 1.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.4 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.7 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 2.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 1.0 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.3 | 1.3 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.3 | 4.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 1.0 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.3 | 4.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.3 | 1.6 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.3 | 5.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 1.0 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 1.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.9 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.3 | 2.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 1.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 6.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 1.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 0.9 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 8.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.3 | 0.8 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 1.4 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.3 | 8.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 1.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 1.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.3 | 3.2 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.3 | 2.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.3 | 4.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 3.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 1.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 6.0 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.3 | 1.0 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 1.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 0.7 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 1.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.2 | 3.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.2 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.2 | 4.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 0.9 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 3.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 1.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 4.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.2 | 0.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 0.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 2.0 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 1.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 1.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 4.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 7.3 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.2 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 3.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 0.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.2 | 13.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 1.5 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.2 | 3.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 3.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 5.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.2 | 2.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 0.8 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.2 | 0.4 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.2 | 1.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 1.6 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.2 | 1.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 1.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.2 | 5.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 4.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 3.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.2 | 0.6 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.2 | 3.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 2.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 1.3 | GO:1904375 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.2 | 0.6 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 2.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 3.8 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.2 | 1.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.7 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.2 | 2.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 2.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 3.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 3.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.5 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.5 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 1.7 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 2.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 1.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.2 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 3.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 3.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 2.9 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.2 | 1.7 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 2.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 2.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.2 | 3.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 12.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.2 | 2.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.7 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 3.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.5 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 1.3 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 4.9 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 4.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.9 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 | 2.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.4 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.1 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.1 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.4 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.1 | 2.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 2.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.4 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.1 | 3.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 4.7 | GO:1903052 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.1 | 0.4 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.1 | 1.0 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 1.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.5 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 4.9 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 1.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.9 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.1 | 0.6 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 2.0 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.5 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.9 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.3 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 1.0 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.4 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.1 | 1.5 | GO:2000223 | posterior lateral line neuromast deposition(GO:0048922) regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 2.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 3.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 1.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.5 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.1 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 5.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.3 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.1 | 3.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.4 | GO:0090280 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 4.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.6 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.8 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 3.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 3.1 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.1 | 4.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 2.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 3.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 1.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.5 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.7 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.1 | 0.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.7 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.1 | 0.6 | GO:0045923 | positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.1 | 1.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 6.0 | GO:0016485 | protein processing(GO:0016485) |
| 0.1 | 0.7 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.4 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.5 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.1 | 5.3 | GO:1903706 | regulation of hemopoiesis(GO:1903706) |
| 0.1 | 3.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 4.3 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 2.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.3 | GO:0072425 | G2 DNA damage checkpoint(GO:0031572) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 1.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.1 | 24.7 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.0 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 0.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 1.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.7 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 2.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 21.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 1.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.5 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 3.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 2.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 3.1 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.2 | GO:0045862 | positive regulation of proteolysis(GO:0045862) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.3 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 3.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) DNA strand elongation(GO:0022616) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 1.1 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.5 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.5 | GO:0010766 | positive regulation of sodium ion transport(GO:0010765) negative regulation of sodium ion transport(GO:0010766) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.1 | GO:0030098 | lymphocyte differentiation(GO:0030098) |
| 0.0 | 1.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.4 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0032366 | intracellular lipid transport(GO:0032365) intracellular sterol transport(GO:0032366) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 1.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.0 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 3.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 5.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.5 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 0.6 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.4 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.8 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 1.1 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.0 | GO:0033005 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of B cell mediated immunity(GO:0002712) positive regulation of B cell mediated immunity(GO:0002714) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) regulation of immunoglobulin mediated immune response(GO:0002889) positive regulation of immunoglobulin mediated immune response(GO:0002891) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.3 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.0 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.1 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0071621 | neutrophil chemotaxis(GO:0030593) granulocyte chemotaxis(GO:0071621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.3 | 3.9 | GO:0098536 | deuterosome(GO:0098536) |
| 1.0 | 5.0 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.9 | 3.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.9 | 3.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.8 | 4.6 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.7 | 8.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.6 | 1.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.6 | 3.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 3.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.6 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 2.8 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 8.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.6 | 3.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 2.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 3.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.5 | 3.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.5 | 10.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.5 | 4.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.5 | 2.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 9.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 2.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 1.7 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.4 | 5.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 1.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 7.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.4 | 8.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 2.8 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.3 | 3.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.3 | 2.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 3.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 2.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.3 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 4.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 1.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 2.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 1.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 1.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 2.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 2.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 6.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 5.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 3.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 4.5 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 2.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 1.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 3.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 3.0 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 4.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 0.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 2.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 0.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 2.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 1.2 | GO:0030684 | preribosome(GO:0030684) |
| 0.2 | 4.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 5.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 2.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 3.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.4 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 6.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 8.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 13.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 7.8 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.1 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 26.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 6.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 8.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 4.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 4.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 11.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 6.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 6.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 1.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 2.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 4.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.2 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.1 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 13.5 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 5.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 3.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 4.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 4.9 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.9 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 8.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 5.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 19.5 | GO:0031981 | nuclear lumen(GO:0031981) |
| 0.0 | 6.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 6.9 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 102.8 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.2 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.3 | GO:0005938 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
| 0.0 | 1.1 | GO:0019867 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0022624 | proteasome regulatory particle(GO:0005838) proteasome accessory complex(GO:0022624) |
| 0.0 | 0.9 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.6 | 6.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 1.4 | 4.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.3 | 5.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 1.3 | 10.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 1.3 | 5.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.1 | 7.9 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.1 | 3.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.0 | 14.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.9 | 6.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.8 | 2.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.7 | 2.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.7 | 2.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.6 | 5.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.6 | 2.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.6 | 2.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.6 | 2.8 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.6 | 2.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.5 | 0.5 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.5 | 5.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 2.1 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.5 | 3.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.5 | 3.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 1.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.5 | 5.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 1.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 5.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.5 | 4.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.4 | 4.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.4 | 4.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 1.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 1.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.4 | 2.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 1.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 3.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 1.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 6.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 1.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.3 | 4.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 3.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 2.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.3 | 3.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 1.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 3.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 0.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 4.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 5.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 2.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.3 | 2.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 0.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 3.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 1.4 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.3 | 3.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.3 | 0.8 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 0.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 1.3 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.3 | 6.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 1.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 18.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.2 | 1.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 4.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 3.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 1.0 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.2 | 3.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.2 | 3.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 2.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 1.2 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 0.6 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 3.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 1.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 4.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 2.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.7 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.2 | 4.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.5 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.2 | 1.5 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.2 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 2.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 0.8 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 1.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.8 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 4.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 0.6 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.5 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.7 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 3.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 2.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 2.2 | GO:0004532 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 5.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 1.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 36.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.7 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 16.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 9.9 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 4.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 1.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 4.6 | GO:0043130 | ubiquitin-like protein binding(GO:0032182) ubiquitin binding(GO:0043130) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 14.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 2.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 7.9 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 7.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 18.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 4.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 1.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 7.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.7 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 2.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 5.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 3.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.0 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 14.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 2.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 7.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 12.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 1.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 15.4 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0061133 | peptidase activator activity(GO:0016504) endopeptidase activator activity(GO:0061133) |
| 0.0 | 3.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 12.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 5.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 11.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.5 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 4.1 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 1.4 | GO:0030898 | microfilament motor activity(GO:0000146) actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 2.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.5 | 2.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 2.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 10.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 2.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 11.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 3.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 4.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 9.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 5.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 8.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.4 | 9.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.4 | 9.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.4 | 1.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 4.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 5.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 4.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.3 | 5.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 3.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 3.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 4.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 1.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 2.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 2.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 9.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 7.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 1.6 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.2 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.2 | 5.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 8.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 11.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 5.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 5.8 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 2.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 5.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 13.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.2 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 4.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |