Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
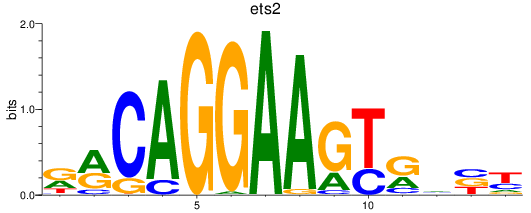
Results for ets2
Z-value: 1.23
Transcription factors associated with ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ets2
|
ENSDARG00000103980 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ets2 | dr11_v1_chr10_+_187760_187764 | 0.51 | 1.6e-01 | Click! |
Activity profile of ets2 motif
Sorted Z-values of ets2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50135492 | 2.00 |
ENSDART00000149398
|
si:ch73-132k15.2
|
si:ch73-132k15.2 |
| chr12_-_4301234 | 1.99 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr12_-_42368296 | 1.77 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr11_+_18037729 | 1.68 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr7_+_24153070 | 1.64 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr20_-_164300 | 1.62 |
ENSDART00000183354
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr20_+_54312970 | 1.60 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr20_+_27087539 | 1.59 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr20_+_54304800 | 1.59 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr14_-_897874 | 1.52 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr18_-_11595567 | 1.52 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr11_+_18175893 | 1.49 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr20_+_54299419 | 1.48 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr11_+_18183220 | 1.47 |
ENSDART00000113468
|
LO018315.10
|
|
| chr5_+_40835601 | 1.46 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr20_+_54309148 | 1.45 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr20_+_54290356 | 1.43 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr11_+_18053333 | 1.43 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr4_-_17263210 | 1.43 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr11_+_18157260 | 1.41 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr19_+_26340736 | 1.41 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr3_-_2613990 | 1.38 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr11_+_18130300 | 1.34 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr20_+_54295213 | 1.34 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr10_+_40324395 | 1.29 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr19_-_29887629 | 1.28 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr8_+_8643901 | 1.23 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr19_+_43579786 | 1.20 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr7_+_13830052 | 1.18 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr15_+_34939906 | 1.15 |
ENSDART00000131182
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr20_+_23947004 | 1.15 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr15_+_34940098 | 1.14 |
ENSDART00000099703
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr7_-_69429561 | 1.13 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr5_+_40837539 | 1.12 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr18_+_15271993 | 1.11 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr9_-_32158288 | 1.09 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr13_-_35459928 | 1.07 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr1_+_59314675 | 1.05 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr3_+_44062576 | 1.03 |
ENSDART00000161277
ENSDART00000168784 |
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr18_-_370286 | 1.01 |
ENSDART00000162633
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr24_+_8904135 | 1.00 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr13_-_25408387 | 1.00 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr23_+_19594608 | 0.99 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr2_-_31711547 | 0.99 |
ENSDART00000175525
ENSDART00000189763 ENSDART00000113498 |
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr18_+_27598755 | 0.98 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr16_+_51462620 | 0.97 |
ENSDART00000169443
|
SLC9A1
|
solute carrier family 9 member A1 |
| chr19_+_43523690 | 0.97 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr2_+_50094873 | 0.97 |
ENSDART00000132307
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr15_+_34946779 | 0.96 |
ENSDART00000192661
ENSDART00000188800 ENSDART00000156515 |
si:ch73-95l15.5
zgc:55621
|
si:ch73-95l15.5 zgc:55621 |
| chr22_-_22337382 | 0.96 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr8_+_41035230 | 0.95 |
ENSDART00000141349
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr16_+_47207691 | 0.94 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr4_-_12781182 | 0.93 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr6_+_2190214 | 0.93 |
ENSDART00000156716
|
acvr1bb
|
activin A receptor type 1Bb |
| chr14_+_24845941 | 0.93 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr16_-_25233515 | 0.91 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr5_-_16475682 | 0.91 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr7_-_30605089 | 0.90 |
ENSDART00000173775
ENSDART00000173789 ENSDART00000166046 ENSDART00000111626 |
rnf111
|
ring finger protein 111 |
| chr16_+_13965923 | 0.90 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr2_+_37207461 | 0.89 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr13_-_34858500 | 0.89 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr14_-_16807206 | 0.89 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr22_-_16758438 | 0.87 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr7_+_73295890 | 0.87 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr17_-_5610514 | 0.86 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr10_-_25543227 | 0.86 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr22_-_38274188 | 0.86 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_-_36364903 | 0.86 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr20_+_4157815 | 0.86 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr7_+_59212666 | 0.85 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr15_+_34934568 | 0.85 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr6_+_54498220 | 0.85 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr25_+_5015019 | 0.84 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr20_-_34028967 | 0.84 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr6_-_15065376 | 0.84 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr21_-_22117085 | 0.83 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr7_+_24528430 | 0.83 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr13_-_17860307 | 0.83 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr14_-_48765262 | 0.82 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr21_+_3941758 | 0.82 |
ENSDART00000181345
|
setx
|
senataxin |
| chr18_+_18405992 | 0.81 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr7_+_13824150 | 0.81 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr12_+_28955766 | 0.81 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr1_-_14258409 | 0.81 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr14_+_16083818 | 0.81 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr21_-_5056812 | 0.81 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr10_-_13396720 | 0.80 |
ENSDART00000030976
|
il11ra
|
interleukin 11 receptor, alpha |
| chr2_-_32592084 | 0.80 |
ENSDART00000184752
|
fastk
|
Fas-activated serine/threonine kinase |
| chr11_-_3535537 | 0.80 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr1_+_24482406 | 0.79 |
ENSDART00000163140
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr13_-_22961605 | 0.79 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr24_-_2423791 | 0.79 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr17_+_5931530 | 0.79 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr12_+_47794089 | 0.78 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr25_+_35889102 | 0.78 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr20_+_16173618 | 0.77 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr5_-_68779747 | 0.77 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr2_+_24700922 | 0.77 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr19_-_24757231 | 0.77 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr20_-_24183333 | 0.77 |
ENSDART00000025862
ENSDART00000153075 |
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr12_-_17810543 | 0.76 |
ENSDART00000090484
|
tecpr1a
|
tectonin beta-propeller repeat containing 1a |
| chr16_-_46393154 | 0.76 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr1_+_44053641 | 0.76 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr6_+_40554551 | 0.76 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr19_+_24891747 | 0.75 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr20_+_29587995 | 0.75 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr10_-_8046764 | 0.75 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr8_+_21254192 | 0.74 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr7_+_26545502 | 0.73 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr5_+_6672870 | 0.73 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr11_+_24314148 | 0.73 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr24_-_1985007 | 0.72 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr7_+_24528866 | 0.72 |
ENSDART00000180552
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr8_+_15277874 | 0.72 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr15_+_12436220 | 0.72 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr7_-_53117131 | 0.71 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr20_+_51813432 | 0.71 |
ENSDART00000127444
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr3_-_21061931 | 0.70 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr8_+_8947623 | 0.70 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr21_+_15790366 | 0.69 |
ENSDART00000101956
|
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr22_+_34784075 | 0.69 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr20_-_1268863 | 0.69 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr5_+_17780475 | 0.69 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr19_-_15192638 | 0.69 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr1_-_55044256 | 0.69 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr10_+_29771256 | 0.68 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr10_+_15608326 | 0.68 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr1_-_44048798 | 0.68 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr1_-_24349759 | 0.67 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr20_-_40360571 | 0.67 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_24313931 | 0.67 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr21_-_12749501 | 0.66 |
ENSDART00000179724
|
LO018011.1
|
|
| chr5_-_54712159 | 0.66 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr6_-_39631164 | 0.66 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr2_-_51087077 | 0.66 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr8_-_45939691 | 0.65 |
ENSDART00000040066
ENSDART00000132297 |
adam9
|
ADAM metallopeptidase domain 9 |
| chr25_+_1335530 | 0.65 |
ENSDART00000090803
|
fem1b
|
fem-1 homolog b (C. elegans) |
| chr22_-_16377666 | 0.65 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr2_-_21167652 | 0.64 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr3_-_32831971 | 0.64 |
ENSDART00000075270
|
zgc:153733
|
zgc:153733 |
| chr1_-_48933 | 0.64 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr6_+_2195625 | 0.64 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr23_-_19682971 | 0.64 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr19_-_3821678 | 0.64 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr25_-_25575717 | 0.64 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr6_-_53204808 | 0.64 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr18_-_399554 | 0.63 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr11_-_18283886 | 0.63 |
ENSDART00000019248
|
stimate
|
STIM activating enhance |
| chr22_-_193234 | 0.63 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr20_-_53078607 | 0.63 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr6_-_42949184 | 0.63 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr13_-_33321058 | 0.63 |
ENSDART00000112084
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr13_+_28580357 | 0.63 |
ENSDART00000007211
|
wbp1la
|
WW domain binding protein 1-like a |
| chr19_-_5805923 | 0.62 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr5_+_27267186 | 0.62 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr15_-_34934784 | 0.62 |
ENSDART00000190848
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr8_-_52594111 | 0.62 |
ENSDART00000167667
|
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr1_+_46405294 | 0.62 |
ENSDART00000143228
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr20_-_4031475 | 0.62 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr6_-_46735380 | 0.61 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr5_-_32336613 | 0.61 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr11_+_13176568 | 0.60 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr22_-_16377960 | 0.60 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr2_+_27651984 | 0.60 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr2_-_20715094 | 0.60 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr11_+_36777613 | 0.60 |
ENSDART00000065644
ENSDART00000158151 |
parapinopsinb
|
parapinopsin b |
| chr17_-_11418513 | 0.60 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_34237189 | 0.60 |
ENSDART00000179624
|
tipin
|
timeless interacting protein |
| chr1_+_494297 | 0.60 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr3_+_17456428 | 0.59 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_+_36713477 | 0.59 |
ENSDART00000181105
ENSDART00000161897 ENSDART00000189952 |
abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr10_-_8053753 | 0.59 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr25_+_4855549 | 0.59 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr21_-_41369539 | 0.59 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr25_-_25575241 | 0.59 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr25_-_37186894 | 0.58 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr13_-_4134141 | 0.58 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr25_+_2361721 | 0.58 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr20_+_22130284 | 0.57 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr3_-_62417677 | 0.57 |
ENSDART00000101888
|
sstr2b
|
somatostatin receptor 2b |
| chr10_+_2582254 | 0.57 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr8_+_36500061 | 0.57 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr19_-_15192840 | 0.57 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr25_-_3745393 | 0.57 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr20_-_26060154 | 0.56 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr10_-_8053385 | 0.56 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr11_+_6882362 | 0.56 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr22_-_20403194 | 0.56 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr21_+_26748141 | 0.56 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr16_+_38201840 | 0.56 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr23_-_36884012 | 0.55 |
ENSDART00000137282
|
ube2j2
|
ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
| chr24_+_8904741 | 0.55 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr7_+_33372680 | 0.54 |
ENSDART00000193436
ENSDART00000099988 |
glceb
|
glucuronic acid epimerase b |
| chr5_+_50953240 | 0.54 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr14_-_40797117 | 0.54 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr24_+_25822999 | 0.54 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr22_+_20710434 | 0.54 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr19_-_34117056 | 0.54 |
ENSDART00000158677
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr8_-_14554785 | 0.54 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr2_-_37281748 | 0.54 |
ENSDART00000137598
|
nadkb
|
NAD kinase b |
Network of associatons between targets according to the STRING database.
First level regulatory network of ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.4 | 2.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.4 | 1.1 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.3 | 1.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 0.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 0.9 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 1.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.3 | 0.8 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 1.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.2 | 4.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 0.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 1.0 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.9 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.2 | 1.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.6 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.2 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.9 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.5 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 4.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.6 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.5 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.5 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.6 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.4 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.8 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 1.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.6 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.3 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.2 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 1.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.6 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.9 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.8 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.3 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.5 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.6 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.3 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 2.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.3 | GO:1901799 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.1 | 0.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.9 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.7 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0072677 | monocyte chemotaxis(GO:0002548) eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 1.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.3 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 3.1 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.4 | GO:0051443 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 1.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 1.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.7 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 1.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0051597 | release of cytochrome c from mitochondria(GO:0001836) response to methylmercury(GO:0051597) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.0 | GO:0002902 | B cell apoptotic process(GO:0001783) tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) regulation of leukocyte apoptotic process(GO:2000106) positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 | 0.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.7 | GO:0071241 | cellular response to inorganic substance(GO:0071241) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 1.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.4 | 1.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 0.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 0.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.5 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.5 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 1.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.7 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 3.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.9 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 1.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.3 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 0.9 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 0.8 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 1.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.3 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 0.8 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 2.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 2.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 2.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.5 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 1.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.6 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.5 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.5 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.0 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 2.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.5 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 7.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 1.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |