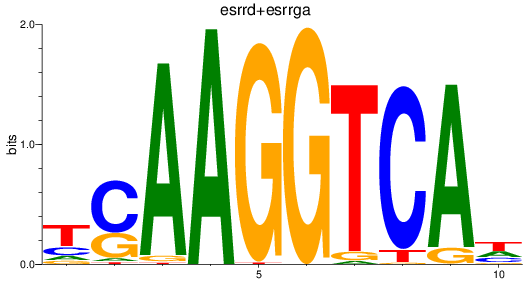
Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for esrrd+esrrga_esrrb_esrra+esrrgb
Z-value: 1.78
Transcription factors associated with esrrd+esrrga_esrrb_esrra+esrrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esrrga
|
ENSDARG00000004861 | estrogen-related receptor gamma a |
|
esrrd
|
ENSDARG00000015064 | estrogen-related receptor delta |
|
esrrb
|
ENSDARG00000100847 | estrogen-related receptor beta |
|
esrrgb
|
ENSDARG00000011696 | estrogen-related receptor gamma b |
|
esrra
|
ENSDARG00000069266 | estrogen-related receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esrrgb | dr11_v1_chr20_-_46817223_46817223 | 0.91 | 7.8e-04 | Click! |
| esrrb | dr11_v1_chr17_+_52300018_52300018 | 0.90 | 9.5e-04 | Click! |
| esrra | dr11_v1_chr21_+_26612777_26612777 | -0.29 | 4.6e-01 | Click! |
| esrrd | dr11_v1_chr18_-_48992363_48992363 | -0.23 | 5.6e-01 | Click! |
Activity profile of esrrd+esrrga_esrrb_esrra+esrrgb motif
Sorted Z-values of esrrd+esrrga_esrrb_esrra+esrrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_45338073 | 3.42 |
ENSDART00000185024
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr14_-_35892767 | 3.19 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr22_+_25720725 | 2.47 |
ENSDART00000150778
|
si:dkeyp-98a7.8
|
si:dkeyp-98a7.8 |
| chr5_-_37103487 | 2.44 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_45250924 | 2.33 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr2_+_30032303 | 2.19 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr1_-_55262763 | 2.18 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr22_+_34784075 | 2.12 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr16_+_50741154 | 2.08 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr6_-_4214297 | 1.95 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr16_-_46578523 | 1.95 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr25_+_5604512 | 1.91 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr12_+_20691310 | 1.91 |
ENSDART00000064335
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr3_-_34724879 | 1.84 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr5_+_37406358 | 1.82 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr21_+_12056934 | 1.71 |
ENSDART00000125380
|
zgc:162344
|
zgc:162344 |
| chr1_-_23110740 | 1.71 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr5_-_57820873 | 1.69 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr5_-_67115872 | 1.63 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr9_+_29548195 | 1.60 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr5_-_57879138 | 1.60 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr13_-_15082024 | 1.58 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr15_+_45994123 | 1.54 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr6_+_2190214 | 1.52 |
ENSDART00000156716
|
acvr1bb
|
activin A receptor type 1Bb |
| chr16_-_30655980 | 1.50 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr13_+_15800742 | 1.43 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr6_+_46481024 | 1.42 |
ENSDART00000155596
|
si:dkey-7k24.5
|
si:dkey-7k24.5 |
| chr19_-_5103313 | 1.41 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_21150845 | 1.40 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr11_+_18175893 | 1.40 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr7_-_48805181 | 1.39 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr17_-_38887424 | 1.38 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr19_+_7636941 | 1.38 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr19_-_5103141 | 1.37 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr4_-_20235904 | 1.37 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr7_+_48297842 | 1.35 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr12_-_45304971 | 1.33 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr13_-_4018888 | 1.31 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr21_-_28640316 | 1.29 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr17_-_1703259 | 1.25 |
ENSDART00000156489
|
xgb
|
x globin |
| chr13_+_30572172 | 1.25 |
ENSDART00000010052
ENSDART00000144417 |
ppifa
|
peptidylprolyl isomerase Fa |
| chr18_+_44769027 | 1.24 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr21_+_13383413 | 1.23 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr10_-_19011934 | 1.21 |
ENSDART00000167973
|
adra1ab
|
adrenoceptor alpha 1Ab |
| chr3_+_3454610 | 1.18 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr11_+_18205766 | 1.17 |
ENSDART00000175559
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr10_+_1849874 | 1.17 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr24_-_20641000 | 1.15 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr23_+_24272421 | 1.15 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr23_-_20002459 | 1.15 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr15_-_7598294 | 1.14 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr23_-_14918276 | 1.14 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr3_-_21166597 | 1.13 |
ENSDART00000175941
|
taok2a
|
TAO kinase 2a |
| chr11_+_18037729 | 1.11 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr7_+_38716048 | 1.11 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr11_+_15890984 | 1.10 |
ENSDART00000158433
|
pank4
|
pantothenate kinase 4 |
| chr18_-_26510545 | 1.07 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr5_+_37729207 | 1.07 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr17_-_30521043 | 1.07 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr2_+_32743807 | 1.06 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr23_+_27756984 | 1.04 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr24_+_37080771 | 1.04 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr1_+_36772348 | 1.03 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr16_-_35427060 | 1.03 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr15_-_7598542 | 1.01 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr11_+_18053333 | 1.01 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr3_+_42923275 | 1.00 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr17_-_45254585 | 0.99 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr24_-_1657276 | 0.99 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr8_+_52530889 | 0.99 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr20_+_16170848 | 0.99 |
ENSDART00000182115
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr1_+_24557414 | 0.98 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr25_+_3104959 | 0.98 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr2_+_24700922 | 0.98 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr11_-_18791834 | 0.97 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr8_+_26879358 | 0.97 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr5_-_16405651 | 0.96 |
ENSDART00000163942
|
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr14_-_16807206 | 0.95 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr23_-_27050083 | 0.94 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr7_-_50633285 | 0.94 |
ENSDART00000174018
|
crtc3
|
CREB regulated transcription coactivator 3 |
| chr23_+_16469530 | 0.94 |
ENSDART00000132898
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr17_+_23554932 | 0.92 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr1_-_55248496 | 0.91 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr14_+_1240419 | 0.91 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr15_-_2841677 | 0.90 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr17_+_21546993 | 0.90 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr9_-_27717006 | 0.89 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr19_-_42045372 | 0.89 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr10_-_18463934 | 0.89 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr10_+_17235370 | 0.88 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr5_+_61738276 | 0.88 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr14_+_30340251 | 0.88 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr1_-_54718863 | 0.87 |
ENSDART00000122601
|
pgam1b
|
phosphoglycerate mutase 1b |
| chr9_-_29844596 | 0.87 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr11_+_18130300 | 0.86 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr11_+_18157260 | 0.85 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr12_-_33359052 | 0.84 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr13_+_33462232 | 0.84 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr19_-_5805923 | 0.84 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr7_+_40094081 | 0.83 |
ENSDART00000186054
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr22_-_38258053 | 0.82 |
ENSDART00000132516
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_-_60142530 | 0.82 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr3_-_21280373 | 0.82 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr22_+_33362552 | 0.81 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr7_+_24523017 | 0.81 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr10_-_5814101 | 0.80 |
ENSDART00000161903
|
il6st
|
interleukin 6 signal transducer |
| chr14_+_32852388 | 0.80 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr24_+_14240196 | 0.79 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr2_-_55298075 | 0.78 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr7_-_60351537 | 0.78 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr4_-_8060962 | 0.78 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr21_+_3960583 | 0.77 |
ENSDART00000149788
|
setx
|
senataxin |
| chr21_+_39432248 | 0.77 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr11_+_31380495 | 0.77 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr23_+_2714949 | 0.76 |
ENSDART00000105284
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr16_+_16266428 | 0.76 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr23_+_44611864 | 0.76 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr14_+_35414632 | 0.76 |
ENSDART00000191516
ENSDART00000084914 |
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr22_-_31517300 | 0.76 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr10_+_8437930 | 0.76 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr15_-_34865952 | 0.75 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr15_+_20801253 | 0.75 |
ENSDART00000179387
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr10_-_38443312 | 0.75 |
ENSDART00000141260
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr14_+_1240235 | 0.75 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr24_+_8904135 | 0.74 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr8_+_37749263 | 0.74 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr8_-_49908978 | 0.73 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr5_+_33301005 | 0.73 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr24_+_33802528 | 0.72 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr21_-_41369539 | 0.72 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr7_+_32605705 | 0.72 |
ENSDART00000173841
|
fshb
|
follicle stimulating hormone, beta polypeptide |
| chr2_-_37352514 | 0.71 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr16_-_33095161 | 0.71 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr5_-_1274794 | 0.70 |
ENSDART00000167337
ENSDART00000125247 |
ciz1a
|
cdkn1a interacting zinc finger protein 1a |
| chr14_-_46113321 | 0.70 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr23_+_16430559 | 0.69 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr7_+_35193832 | 0.69 |
ENSDART00000189002
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr18_+_910992 | 0.69 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr21_+_39197628 | 0.68 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr12_+_28799988 | 0.68 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr15_+_44472356 | 0.68 |
ENSDART00000155763
ENSDART00000028011 |
msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr6_+_19948043 | 0.68 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr2_+_34767171 | 0.67 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr9_+_22003942 | 0.67 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr3_+_4997545 | 0.67 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr6_-_40446536 | 0.66 |
ENSDART00000153466
|
tatdn2
|
TatD DNase domain containing 2 |
| chr1_-_58868306 | 0.66 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr10_-_39281475 | 0.66 |
ENSDART00000175136
|
cry5
|
cryptochrome circadian clock 5 |
| chr2_-_15040345 | 0.66 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr15_+_19990068 | 0.66 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr10_+_44584614 | 0.65 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr13_-_20540790 | 0.65 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr1_+_16574312 | 0.64 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr16_-_2504005 | 0.64 |
ENSDART00000089944
|
wdr26a
|
WD repeat domain 26a |
| chr9_+_13638329 | 0.63 |
ENSDART00000143432
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr6_+_28428329 | 0.63 |
ENSDART00000188056
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_+_19281189 | 0.63 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr4_+_11723852 | 0.63 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr22_+_29113796 | 0.63 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr6_+_19933763 | 0.62 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr14_-_31465905 | 0.62 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr24_-_10394277 | 0.61 |
ENSDART00000127568
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr5_+_35786141 | 0.61 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr14_-_14604527 | 0.60 |
ENSDART00000190378
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr5_-_38064065 | 0.60 |
ENSDART00000137181
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr10_-_25217347 | 0.60 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr1_+_40613297 | 0.60 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr18_-_39636348 | 0.59 |
ENSDART00000129828
|
cyp19a1a
|
cytochrome P450, family 19, subfamily A, polypeptide 1a |
| chr6_-_43283122 | 0.59 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr21_+_33311622 | 0.59 |
ENSDART00000163808
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr2_+_53359234 | 0.59 |
ENSDART00000147581
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr23_+_39346774 | 0.59 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr7_+_7027199 | 0.59 |
ENSDART00000193180
|
rbm14b
|
RNA binding motif protein 14b |
| chr5_+_6672870 | 0.59 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr1_+_17527342 | 0.57 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr13_+_7387822 | 0.57 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr2_+_26060528 | 0.57 |
ENSDART00000058111
|
grin3ba
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Ba |
| chr1_+_44826367 | 0.57 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr25_-_21492630 | 0.57 |
ENSDART00000141481
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr25_+_16214854 | 0.57 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr3_+_60589157 | 0.57 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr21_+_27513859 | 0.57 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr24_+_32472155 | 0.56 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr11_+_13071645 | 0.55 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr14_+_1240604 | 0.55 |
ENSDART00000188258
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr19_-_30510259 | 0.55 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr21_-_41369370 | 0.55 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr11_-_43200994 | 0.55 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr2_-_57076687 | 0.54 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr2_-_37312927 | 0.54 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr9_-_18735256 | 0.54 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr8_-_49495584 | 0.53 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr11_-_42230491 | 0.53 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr9_+_9927516 | 0.53 |
ENSDART00000161089
ENSDART00000168559 |
ttf2
|
transcription termination factor, RNA polymerase II |
| chr20_+_37794633 | 0.53 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr5_+_66132394 | 0.53 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr14_+_35892802 | 0.53 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr24_-_1021318 | 0.53 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr5_-_5669879 | 0.52 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr19_+_40071561 | 0.52 |
ENSDART00000191109
|
zmym4
|
zinc finger, MYM-type 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of esrrd+esrrga_esrrb_esrra+esrrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.7 | 2.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 2.0 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.3 | 1.0 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.3 | 1.2 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.3 | 0.9 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 0.9 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.3 | 1.5 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 0.9 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.3 | 0.9 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.3 | 1.2 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.3 | 1.6 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.3 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.4 | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) |
| 0.2 | 0.6 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.2 | 0.8 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.3 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.2 | 1.8 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 2.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 0.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 1.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.2 | 1.6 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 2.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 2.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.0 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.8 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 1.1 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 0.7 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.1 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.4 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.5 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.3 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.7 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.6 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.2 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 1.0 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.5 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.2 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.4 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.0 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 2.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 5.2 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 1.1 | GO:0046364 | hexose biosynthetic process(GO:0019319) monosaccharide biosynthetic process(GO:0046364) |
| 0.0 | 0.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 2.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 1.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.0 | GO:0010677 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.5 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 1.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 1.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 1.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.4 | GO:0034284 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to monosaccharide(GO:0034284) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0044060 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.6 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.3 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 1.9 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0032869 | response to insulin(GO:0032868) cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:1903313 | positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 2.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.8 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.0 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) regulation of delayed rectifier potassium channel activity(GO:1902259) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.9 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.8 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 0.9 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 3.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 3.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 3.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 2.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.4 | 1.2 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 2.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 1.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 1.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.3 | 1.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 0.9 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.3 | 1.6 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.3 | 0.8 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 3.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.7 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.2 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 2.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.5 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 2.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.9 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 1.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 2.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 3.1 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 2.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.2 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 1.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0016885 | CoA carboxylase activity(GO:0016421) ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.7 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.9 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 2.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 2.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 3.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 1.1 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |