Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
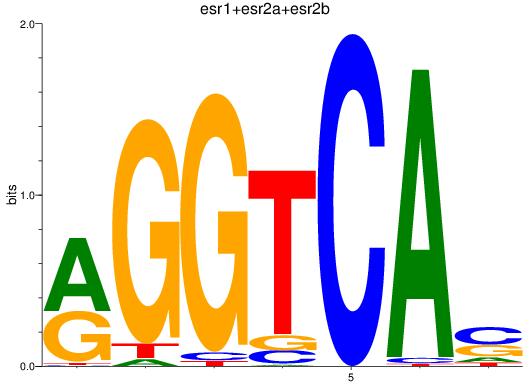
Results for esr1+esr2a+esr2b
Z-value: 2.50
Transcription factors associated with esr1+esr2a+esr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esr1
|
ENSDARG00000004111 | estrogen receptor 1 |
|
esr2a
|
ENSDARG00000016454 | estrogen receptor 2a |
|
esr2b
|
ENSDARG00000034181 | estrogen receptor 2b |
|
esr1
|
ENSDARG00000112357 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esr1 | dr11_v1_chr20_-_26383368_26383375 | 0.89 | 1.4e-03 | Click! |
| esr2b | dr11_v1_chr13_+_37022601_37022632 | 0.83 | 5.5e-03 | Click! |
| esr2a | dr11_v1_chr20_+_21583639_21583639 | -0.72 | 2.8e-02 | Click! |
Activity profile of esr1+esr2a+esr2b motif
Sorted Z-values of esr1+esr2a+esr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44027391 | 9.55 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr12_-_43685802 | 5.06 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr9_-_22310919 | 4.60 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr5_-_71722257 | 4.07 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr19_-_5332784 | 3.82 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_+_9561066 | 3.66 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr7_+_20017211 | 3.64 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr23_+_44741500 | 3.53 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr9_-_22240052 | 3.44 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr10_-_29900546 | 3.44 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr12_-_4683325 | 3.42 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr9_-_22147567 | 3.40 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr9_-_22272181 | 3.36 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr11_+_30729745 | 3.33 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr9_-_22188117 | 3.31 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr12_-_30846055 | 3.18 |
ENSDART00000075983
|
crygmxl2
|
crystallin, gamma MX, like 2 |
| chr6_+_55032439 | 3.12 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr9_+_54644626 | 3.05 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr9_-_22318511 | 3.04 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr20_-_40717900 | 3.00 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr9_-_22232902 | 2.99 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr5_-_55395964 | 2.97 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr15_+_19797918 | 2.93 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr7_+_40884012 | 2.91 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr3_-_41795917 | 2.91 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr16_-_55028740 | 2.83 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr1_+_17676745 | 2.83 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr16_+_29650698 | 2.81 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr13_-_37127970 | 2.78 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr18_+_62932 | 2.76 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr1_-_59176949 | 2.76 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr11_-_6048490 | 2.75 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr6_-_607063 | 2.75 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr15_+_32711663 | 2.75 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr18_+_31016379 | 2.74 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr22_-_15593824 | 2.69 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr19_+_19759577 | 2.64 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr6_-_13783604 | 2.62 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr2_-_5728843 | 2.60 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr9_-_7539297 | 2.59 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr6_+_15268685 | 2.53 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr2_-_716426 | 2.48 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr7_+_40228422 | 2.39 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr24_-_38374744 | 2.39 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr17_-_6738538 | 2.35 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr3_-_46811611 | 2.30 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr16_+_12022543 | 2.29 |
ENSDART00000012673
|
gnb3a
|
guanine nucleotide binding protein (G protein), beta polypeptide 3a |
| chr14_+_17376940 | 2.29 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr23_-_21471022 | 2.24 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr11_+_7158723 | 2.19 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr3_+_23737795 | 2.18 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr25_+_23591990 | 2.15 |
ENSDART00000151938
ENSDART00000089199 |
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr2_-_24013260 | 2.15 |
ENSDART00000137065
|
col15a1b
|
collagen, type XV, alpha 1b |
| chr4_+_17280868 | 2.14 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr7_-_51032128 | 2.13 |
ENSDART00000182781
ENSDART00000121574 |
col4a6
|
collagen, type IV, alpha 6 |
| chr14_-_17072736 | 2.09 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr5_-_46980651 | 2.08 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr8_-_23758312 | 2.08 |
ENSDART00000132659
|
INAVA
|
si:ch211-163l21.4 |
| chr20_+_54738210 | 2.07 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr7_-_34265481 | 2.07 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr2_+_42191592 | 2.07 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr21_+_40106448 | 2.03 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr9_-_22821901 | 2.01 |
ENSDART00000101711
|
neb
|
nebulin |
| chr4_-_16333944 | 2.00 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr6_-_11768198 | 1.99 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr4_-_191736 | 1.99 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_29096881 | 1.99 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr23_-_26535875 | 1.97 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr1_-_40914752 | 1.97 |
ENSDART00000113087
|
hmx1
|
H6 family homeobox 1 |
| chr17_-_12385308 | 1.95 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr8_+_23213320 | 1.94 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr6_+_12853655 | 1.93 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr20_-_22484621 | 1.93 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr13_+_15004398 | 1.93 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr5_+_1278092 | 1.90 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr15_+_28096152 | 1.89 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr13_-_31435137 | 1.87 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr5_+_20693724 | 1.87 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr5_-_65021736 | 1.85 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr1_+_16144615 | 1.85 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr1_-_12278522 | 1.84 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr5_+_70155935 | 1.83 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr3_-_18575868 | 1.83 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr24_-_39610585 | 1.82 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr16_+_23972126 | 1.82 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr13_-_9525527 | 1.82 |
ENSDART00000190618
|
CR848040.5
|
|
| chr4_-_1360495 | 1.81 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr11_-_24681292 | 1.80 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr16_+_5774977 | 1.80 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr23_-_45705525 | 1.80 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr23_-_46040618 | 1.78 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr20_+_48782068 | 1.75 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr4_+_38344 | 1.75 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr24_-_32408404 | 1.75 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr16_-_560574 | 1.74 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr2_-_31302615 | 1.73 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr23_-_15216654 | 1.72 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr3_+_28953274 | 1.72 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr19_-_30404096 | 1.72 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr19_+_58954 | 1.69 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr22_-_2886937 | 1.69 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr23_-_24488696 | 1.69 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr8_+_46217861 | 1.68 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr3_-_36260102 | 1.68 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr3_-_30941362 | 1.65 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr13_-_36703164 | 1.65 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr1_+_53945934 | 1.64 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr10_+_37145007 | 1.64 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr18_+_2837563 | 1.64 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr16_+_33655890 | 1.63 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr9_-_23033818 | 1.62 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr2_-_48196092 | 1.61 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr15_-_29388012 | 1.61 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr10_+_43994471 | 1.60 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr18_+_48423973 | 1.59 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr8_-_49431939 | 1.58 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr7_+_26716321 | 1.58 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr21_+_27416284 | 1.56 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr19_-_9712530 | 1.54 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr6_-_40058686 | 1.54 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr23_-_31372639 | 1.54 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr18_+_22302635 | 1.53 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr22_+_11756040 | 1.52 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr23_-_39636195 | 1.52 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr18_+_50801307 | 1.51 |
ENSDART00000174126
|
tspan4a
|
tetraspanin 4a |
| chr9_-_1951144 | 1.51 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr10_-_17103651 | 1.50 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr19_+_30662529 | 1.50 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr7_-_26924903 | 1.50 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr8_+_39634114 | 1.50 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr11_+_11201096 | 1.48 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr10_-_31782616 | 1.48 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr18_-_48492951 | 1.48 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr15_-_21165237 | 1.48 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr5_-_41831646 | 1.48 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr6_-_609880 | 1.47 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr8_+_24745041 | 1.47 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr1_+_46194333 | 1.46 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr7_-_4036184 | 1.45 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr15_+_24644016 | 1.43 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr19_-_30403922 | 1.43 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr9_-_710896 | 1.42 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr15_+_5923851 | 1.41 |
ENSDART00000152520
ENSDART00000145827 ENSDART00000121529 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr11_-_30138299 | 1.41 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr10_+_22775253 | 1.41 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr24_-_12938922 | 1.41 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr14_+_36246726 | 1.40 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr10_+_6318227 | 1.39 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr18_-_15373620 | 1.39 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr14_-_215051 | 1.39 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr18_-_16123222 | 1.38 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr15_+_24644251 | 1.38 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr23_+_43954809 | 1.38 |
ENSDART00000164080
|
corin
|
corin, serine peptidase |
| chr6_-_60031693 | 1.38 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr20_-_52314299 | 1.37 |
ENSDART00000038157
|
naprt
|
nicotinate phosphoribosyltransferase |
| chr13_+_22480496 | 1.36 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr5_+_38276582 | 1.36 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr23_-_11870962 | 1.36 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr13_-_39947335 | 1.35 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr16_-_2414063 | 1.34 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr18_-_48517040 | 1.33 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr18_+_30508729 | 1.33 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr6_-_40429411 | 1.32 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr19_+_4916233 | 1.32 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr8_+_48613040 | 1.32 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr10_-_22797959 | 1.32 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr6_+_48618512 | 1.32 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr13_-_16222388 | 1.32 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr1_-_49498116 | 1.31 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr23_-_26536055 | 1.31 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr7_+_26029672 | 1.30 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr1_+_29759678 | 1.30 |
ENSDART00000054059
ENSDART00000101856 |
cpb2
|
carboxypeptidase B2 (plasma) |
| chr23_+_24124684 | 1.30 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr21_+_25688388 | 1.29 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr13_-_31452516 | 1.29 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr1_-_22757145 | 1.29 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr3_+_1182315 | 1.29 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr16_-_38921745 | 1.28 |
ENSDART00000131409
|
BX571839.1
|
|
| chr14_+_33722950 | 1.28 |
ENSDART00000075312
|
apln
|
apelin |
| chr9_-_22834860 | 1.27 |
ENSDART00000146486
|
neb
|
nebulin |
| chr8_-_16697912 | 1.27 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr20_+_2281933 | 1.26 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr6_+_41463786 | 1.26 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr6_+_28124393 | 1.26 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr23_-_45504991 | 1.25 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr2_-_55853943 | 1.25 |
ENSDART00000122576
|
rx2
|
retinal homeobox gene 2 |
| chr23_-_15330168 | 1.24 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr11_+_30295582 | 1.24 |
ENSDART00000122424
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr4_-_16853464 | 1.24 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr20_-_16906623 | 1.24 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr15_-_2652640 | 1.21 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr20_+_40150612 | 1.20 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr2_-_38000276 | 1.20 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr4_-_7212875 | 1.19 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr20_-_24122881 | 1.19 |
ENSDART00000131857
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr8_+_39760258 | 1.19 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr20_+_26349002 | 1.18 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr25_-_15214161 | 1.18 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr10_+_13209580 | 1.18 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr17_+_53425037 | 1.17 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
Network of associatons between targets according to the STRING database.
First level regulatory network of esr1+esr2a+esr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.8 | 2.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.7 | 2.9 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.7 | 2.8 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.6 | 2.6 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.6 | 1.8 | GO:0050995 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.6 | 4.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 3.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.5 | 3.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 1.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.5 | 2.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.5 | 1.5 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.5 | 1.5 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.5 | 1.4 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.5 | 2.3 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.4 | 1.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 | 1.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.4 | 3.5 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.4 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 1.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.4 | 1.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 1.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.4 | 4.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 1.1 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.4 | 1.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.8 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.4 | 46.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.4 | 1.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) response to dexamethasone(GO:0071548) |
| 0.3 | 3.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.3 | 1.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 4.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 1.3 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.3 | 2.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.9 | GO:0033032 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 2.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 1.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 0.9 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 1.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 1.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 0.8 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 0.7 | GO:0035776 | pronephric proximal tubule development(GO:0035776) proximal straight tubule development(GO:0072020) |
| 0.2 | 0.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 0.7 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 0.7 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.7 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.2 | 1.7 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.2 | 0.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.2 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 0.9 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.2 | 1.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 1.7 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 0.9 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 0.6 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 1.3 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 0.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 3.5 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.2 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.2 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 3.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 3.1 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.2 | 0.9 | GO:2000328 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.2 | 2.5 | GO:0014846 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.5 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 0.5 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.5 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 2.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.7 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.2 | 3.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.7 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 1.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 0.6 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 3.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 2.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.5 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 3.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 1.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.8 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 1.7 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.3 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.7 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.2 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 2.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.0 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 3.6 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 2.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 5.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.7 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.0 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.1 | 1.0 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.6 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.9 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 7.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 1.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 1.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.3 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.5 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.7 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 5.0 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 1.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 6.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.6 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 0.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.4 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.5 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.9 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.7 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.1 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0097581 | lamellipodium organization(GO:0097581) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.7 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 2.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 5.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0003272 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 2.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 2.0 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.2 | GO:0035966 | response to topologically incorrect protein(GO:0035966) |
| 0.0 | 1.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 1.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 2.5 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 2.2 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.0 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.5 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 1.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.1 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0043049 | otic placode formation(GO:0043049) ectodermal placode formation(GO:0060788) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 3.0 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 3.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 4.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.8 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 10.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 7.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 3.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 8.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.5 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 20.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 49.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.9 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.7 | 2.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.7 | 3.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.6 | 5.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 47.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 3.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.7 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.4 | 1.3 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.4 | 3.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 1.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 1.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 4.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 1.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 1.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.6 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.3 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.3 | 2.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 4.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 4.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 0.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.3 | 2.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 4.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 2.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 1.8 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 1.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.6 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 1.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 0.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.0 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 1.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 1.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 3.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 4.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 2.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 5.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.7 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.5 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 5.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.4 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.4 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 4.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 4.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 15.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 3.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.6 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 9.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 3.2 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 1.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.7 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 3.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 8.1 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.8 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| 0.0 | 0.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 11.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 1.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 6.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |