Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for en1b_en2a+en2b_gbx1_emx1
Z-value: 0.56
Transcription factors associated with en1b_en2a+en2b_gbx1_emx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
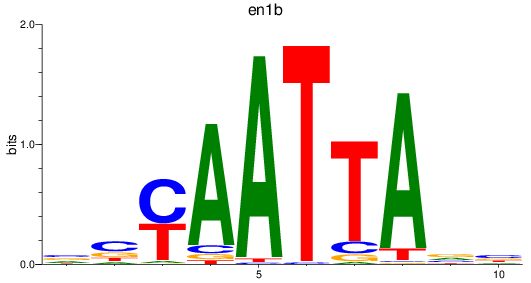
en1b
|
ENSDARG00000098730 | engrailed homeobox 1b |
|
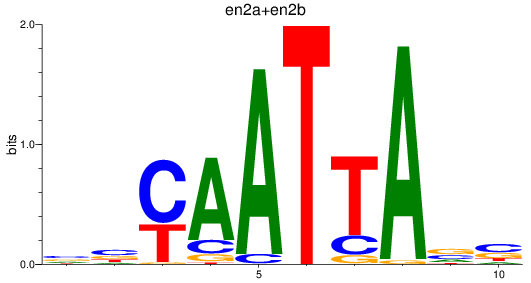
en2a
|
ENSDARG00000026599 | engrailed homeobox 2a |
|
en2b
|
ENSDARG00000038868 | engrailed homeobox 2b |
|
en2a
|
ENSDARG00000115233 | engrailed homeobox 2a |
|
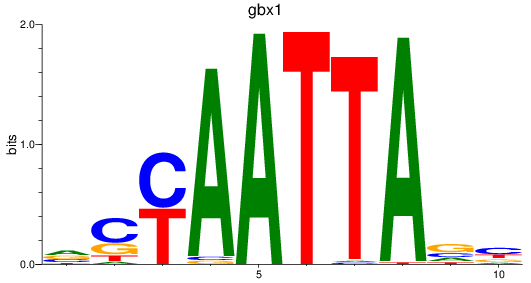
gbx1
|
ENSDARG00000071418 | gastrulation brain homeobox 1 |
|
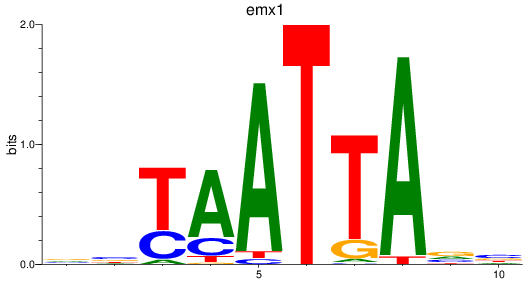
emx1
|
ENSDARG00000039569 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1b | dr11_v1_chr1_+_7679328_7679328 | -0.95 | 6.4e-05 | Click! |
| emx1 | dr11_v1_chr13_+_15004398_15004398 | -0.90 | 1.1e-03 | Click! |
| en2a | dr11_v1_chr7_-_40993456_40993456 | -0.88 | 2.0e-03 | Click! |
| en2b | dr11_v1_chr2_+_29976419_29976419 | -0.83 | 6.1e-03 | Click! |
| gbx1 | dr11_v1_chr24_+_34113424_34113424 | -0.20 | 6.1e-01 | Click! |
Activity profile of en1b_en2a+en2b_gbx1_emx1 motif
Sorted Z-values of en1b_en2a+en2b_gbx1_emx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 2.27 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_21362071 | 1.61 |
ENSDART00000125167
|
avd
|
avidin |
| chr9_-_35633827 | 1.61 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_-_21362320 | 1.60 |
ENSDART00000189789
|
avd
|
avidin |
| chr20_-_23426339 | 1.30 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr10_+_6884627 | 1.18 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr1_-_55248496 | 1.15 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr2_+_6253246 | 1.13 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_+_1023839 | 1.09 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr18_-_40708537 | 1.04 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr10_-_44560165 | 1.01 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr4_+_306036 | 0.97 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr10_-_34002185 | 0.93 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr24_+_12835935 | 0.93 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr11_-_44801968 | 0.89 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr24_-_9979342 | 0.86 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr14_+_34490445 | 0.85 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr19_+_31585917 | 0.82 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr22_+_17828267 | 0.76 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr11_-_1550709 | 0.65 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr10_+_6884123 | 0.64 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr5_-_30074332 | 0.62 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr5_-_15283509 | 0.61 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr22_+_4488454 | 0.60 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr1_-_18811517 | 0.58 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr16_-_42056137 | 0.57 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr8_-_23780334 | 0.56 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr17_+_16046132 | 0.56 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr2_-_15324837 | 0.54 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_+_17371356 | 0.53 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr10_+_19596214 | 0.53 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr17_+_16046314 | 0.51 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_+_37638873 | 0.51 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr9_+_54695981 | 0.51 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr23_+_42254960 | 0.50 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr25_+_35891342 | 0.50 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr5_+_68807170 | 0.49 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr12_-_48168135 | 0.48 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr14_-_8940499 | 0.48 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr12_-_14143344 | 0.46 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr10_-_35257458 | 0.45 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr12_+_46582727 | 0.45 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr2_-_57076687 | 0.45 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr10_-_13343831 | 0.45 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr9_+_18829360 | 0.45 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr8_-_1838315 | 0.45 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr5_-_68779747 | 0.44 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr5_-_68795063 | 0.44 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr3_+_22035863 | 0.43 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr13_-_21660203 | 0.43 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr20_-_6532462 | 0.41 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr24_-_2450597 | 0.41 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr6_-_43283122 | 0.41 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr6_+_28208973 | 0.39 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr18_-_20458840 | 0.39 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr2_-_38287987 | 0.39 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr8_-_53044300 | 0.38 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_+_40922572 | 0.37 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr21_-_31013817 | 0.37 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr23_-_35790235 | 0.36 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr20_+_46513651 | 0.36 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr10_-_32494499 | 0.36 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr2_+_20793982 | 0.35 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr23_+_2728095 | 0.35 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr6_-_12275836 | 0.35 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr16_+_47207691 | 0.35 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr11_+_18183220 | 0.35 |
ENSDART00000113468
|
LO018315.10
|
|
| chr19_-_20403507 | 0.35 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr11_-_6452444 | 0.35 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr2_+_41526904 | 0.34 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr2_+_56213694 | 0.34 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr3_-_23643751 | 0.34 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr18_+_20560616 | 0.33 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr5_+_63302660 | 0.33 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr5_+_6954162 | 0.33 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr17_+_8799451 | 0.32 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr24_+_39518774 | 0.32 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr23_-_17003533 | 0.32 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr22_+_9003090 | 0.32 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr14_-_33945692 | 0.32 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr1_+_35985813 | 0.32 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr9_-_53666031 | 0.32 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr24_-_34680956 | 0.32 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_+_23515966 | 0.31 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr7_-_51368681 | 0.31 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr9_-_3934963 | 0.31 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr19_+_2631565 | 0.30 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr12_-_33357655 | 0.30 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr23_+_27782071 | 0.30 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr23_-_31913069 | 0.30 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_+_34592526 | 0.30 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr19_+_4968947 | 0.29 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr5_-_62925282 | 0.29 |
ENSDART00000188048
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr10_-_32494304 | 0.29 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr6_-_2133737 | 0.29 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr17_+_49500820 | 0.29 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr4_+_11723852 | 0.29 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr7_-_51773166 | 0.28 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr6_+_21001264 | 0.28 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr4_+_25917915 | 0.28 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_32825917 | 0.28 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr3_-_15210491 | 0.27 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr16_+_28994709 | 0.27 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr13_-_35808904 | 0.27 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr17_+_21902058 | 0.27 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr17_-_21162821 | 0.27 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr6_-_37744430 | 0.27 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr25_-_32869794 | 0.26 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr5_+_28497956 | 0.26 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr4_+_13586689 | 0.26 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr8_-_52562672 | 0.26 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr9_-_50000144 | 0.26 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr5_-_19006290 | 0.26 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr7_+_24528430 | 0.26 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr16_-_41714988 | 0.26 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr24_-_10006158 | 0.26 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr19_-_8768564 | 0.25 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr7_-_12464412 | 0.25 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr15_-_16177603 | 0.25 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr24_-_24724233 | 0.25 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr20_-_51831657 | 0.25 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr24_+_40905100 | 0.25 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr19_-_5805923 | 0.25 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr5_-_9625459 | 0.25 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr16_-_41488023 | 0.25 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr20_-_14114078 | 0.24 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr17_-_40956035 | 0.24 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr14_+_26247319 | 0.24 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr3_+_18807006 | 0.24 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr3_+_22443313 | 0.24 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr5_-_66397688 | 0.24 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr8_-_37249813 | 0.24 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr17_-_20118145 | 0.24 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr13_+_38814521 | 0.23 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr11_-_42134968 | 0.23 |
ENSDART00000187115
|
FP325130.1
|
|
| chr15_-_43284021 | 0.23 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr14_+_32942063 | 0.23 |
ENSDART00000187705
|
lnx2b
|
ligand of numb-protein X 2b |
| chr1_-_47071979 | 0.23 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr10_-_33297864 | 0.23 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr20_-_37813863 | 0.23 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_-_67629263 | 0.22 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr24_-_25144441 | 0.22 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_+_12813316 | 0.22 |
ENSDART00000155629
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr12_+_47698356 | 0.22 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr9_+_17983463 | 0.22 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr8_+_49117518 | 0.22 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr7_-_19642417 | 0.21 |
ENSDART00000160936
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr22_-_23267893 | 0.21 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr17_+_24821627 | 0.21 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr19_-_20430892 | 0.21 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr15_-_590787 | 0.21 |
ENSDART00000189367
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr9_+_29548195 | 0.21 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr5_-_68782641 | 0.21 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr24_-_30843250 | 0.21 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr1_-_23308225 | 0.20 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr15_+_28410664 | 0.20 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr23_+_28322986 | 0.20 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr5_-_23715861 | 0.20 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr6_-_40922971 | 0.20 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr8_+_45334255 | 0.20 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_-_51831816 | 0.20 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr8_+_11325310 | 0.20 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr9_-_21460164 | 0.20 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr21_-_15200556 | 0.20 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr6_+_41191482 | 0.20 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr5_-_67241633 | 0.20 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr13_+_22295905 | 0.20 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr17_-_3815693 | 0.20 |
ENSDART00000161207
|
PLCB4
|
phospholipase C beta 4 |
| chr19_-_5865766 | 0.20 |
ENSDART00000191007
|
LO018585.1
|
|
| chr20_+_27087539 | 0.20 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr15_+_17345609 | 0.20 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr3_+_32832538 | 0.20 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr23_+_42272588 | 0.20 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr9_-_38021889 | 0.20 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr3_-_697363 | 0.20 |
ENSDART00000186520
|
dicp1.3-4
|
diverse immunoglobulin domain-containing protein 1.3-4 |
| chr20_-_20324792 | 0.20 |
ENSDART00000042376
|
snapc1a
|
small nuclear RNA activating complex, polypeptide 1a |
| chr19_+_12406583 | 0.20 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr19_-_18313303 | 0.20 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr5_+_19933356 | 0.19 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr20_+_29209926 | 0.19 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr6_-_19665114 | 0.19 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr4_-_63848305 | 0.19 |
ENSDART00000097308
|
BX901974.1
|
|
| chr25_-_27621268 | 0.19 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr18_+_44849809 | 0.19 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr24_+_14581864 | 0.19 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr24_-_10021341 | 0.19 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr13_+_40501455 | 0.19 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr8_-_52909850 | 0.19 |
ENSDART00000161943
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr24_+_19415124 | 0.19 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr13_+_33688474 | 0.18 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr23_-_31913231 | 0.18 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_18031304 | 0.18 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr2_-_21819421 | 0.18 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr11_-_11791718 | 0.18 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr7_+_38089650 | 0.18 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr15_-_4969525 | 0.18 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr2_+_366429 | 0.18 |
ENSDART00000111746
|
si:dkey-33c14.6
|
si:dkey-33c14.6 |
| chr2_+_8779164 | 0.18 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr4_-_77125693 | 0.18 |
ENSDART00000174256
|
CU467646.3
|
|
| chr20_+_54304800 | 0.18 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1b_en2a+en2b_gbx1_emx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.0 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 0.8 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.6 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 2.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 2.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.4 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.5 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.6 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.9 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 1.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.1 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 1.0 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.0 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.0 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.5 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0060281 | regulation of oocyte development(GO:0060281) positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.3 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.0 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.0 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 0.7 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.2 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 3.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 0.5 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 2.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.2 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 2.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 3.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |