Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
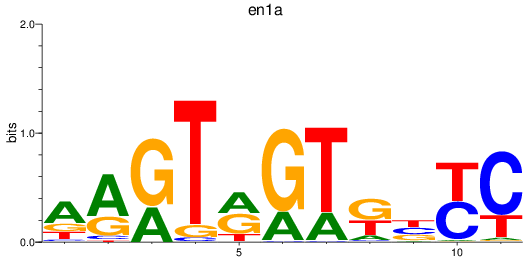
Results for en1a
Z-value: 0.35
Transcription factors associated with en1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en1a
|
ENSDARG00000014321 | engrailed homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1a | dr11_v1_chr9_-_785444_785444 | 0.51 | 1.7e-01 | Click! |
Activity profile of en1a motif
Sorted Z-values of en1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44713135 | 0.70 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr13_-_9525527 | 0.56 |
ENSDART00000190618
|
CR848040.5
|
|
| chr5_-_65021736 | 0.55 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr2_-_16562505 | 0.49 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr19_-_3240605 | 0.47 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr7_+_29954709 | 0.47 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr3_+_26345732 | 0.46 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr3_+_28939759 | 0.42 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr7_+_29955368 | 0.40 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr8_+_13503377 | 0.35 |
ENSDART00000034740
ENSDART00000167187 |
fut9d
|
fucosyltransferase 9d |
| chr23_+_36653376 | 0.33 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr5_-_30615901 | 0.32 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr7_-_34265481 | 0.32 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr14_-_21123551 | 0.31 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr8_-_50147948 | 0.30 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr9_+_21277846 | 0.30 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr7_-_69636502 | 0.30 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr17_-_44249720 | 0.30 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr2_+_26498446 | 0.30 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr5_-_8164439 | 0.29 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr1_-_59287410 | 0.29 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr11_+_3989403 | 0.29 |
ENSDART00000082379
|
spcs1
|
signal peptidase complex subunit 1 |
| chr2_-_24554416 | 0.29 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr19_-_18626515 | 0.28 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr15_-_19771981 | 0.28 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr7_+_31879986 | 0.27 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr24_-_32408404 | 0.27 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr6_+_47424266 | 0.27 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr5_+_69716458 | 0.27 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr23_+_42396934 | 0.27 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr15_-_20233105 | 0.26 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr20_+_16750177 | 0.26 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr5_-_44843738 | 0.25 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr8_+_25616946 | 0.25 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr15_+_23799461 | 0.25 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr20_+_34845672 | 0.25 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr9_-_33121535 | 0.24 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr6_-_13783604 | 0.24 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr20_+_539852 | 0.24 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr4_-_52165969 | 0.23 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr13_-_15700060 | 0.23 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr23_+_42819221 | 0.23 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr20_-_20312789 | 0.23 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr3_+_23768898 | 0.22 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr1_-_13271569 | 0.22 |
ENSDART00000127838
|
pcdh18a
|
protocadherin 18a |
| chr10_+_33982010 | 0.22 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr11_+_23569869 | 0.22 |
ENSDART00000163418
|
si:ch73-21d24.1
|
si:ch73-21d24.1 |
| chr16_+_54780544 | 0.22 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr14_-_9277152 | 0.22 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr1_+_34295925 | 0.21 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr5_-_55625247 | 0.21 |
ENSDART00000133156
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr3_+_14543953 | 0.21 |
ENSDART00000161710
|
epor
|
erythropoietin receptor |
| chr8_+_26565512 | 0.21 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr6_-_39764995 | 0.20 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr19_-_31402429 | 0.20 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr6_+_46320693 | 0.19 |
ENSDART00000128621
ENSDART00000155917 |
si:dkeyp-67f1.2
|
si:dkeyp-67f1.2 |
| chr19_+_43780970 | 0.19 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr12_+_34119439 | 0.19 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr24_+_9372292 | 0.19 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr15_-_2632891 | 0.19 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr17_-_6349044 | 0.19 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr6_-_40195510 | 0.18 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr3_-_6767440 | 0.18 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr23_+_26946744 | 0.18 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr1_-_11913170 | 0.18 |
ENSDART00000165543
|
grk4
|
G protein-coupled receptor kinase 4 |
| chr11_-_44876005 | 0.18 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr18_-_41375120 | 0.18 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr11_+_42633376 | 0.17 |
ENSDART00000076711
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr19_+_25476228 | 0.17 |
ENSDART00000112808
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr2_+_22495274 | 0.17 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr10_+_38512270 | 0.17 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr15_+_37197494 | 0.17 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr2_-_1514001 | 0.17 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr13_-_29424454 | 0.17 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr16_+_5196226 | 0.16 |
ENSDART00000189704
|
soga3a
|
SOGA family member 3a |
| chr3_+_12784460 | 0.16 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr4_-_17805128 | 0.16 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr3_-_41795917 | 0.16 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr6_+_21684296 | 0.16 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr5_-_64203101 | 0.16 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr16_-_45080812 | 0.15 |
ENSDART00000157377
|
si:rp71-77l1.2
|
si:rp71-77l1.2 |
| chr1_+_54865552 | 0.15 |
ENSDART00000145381
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr15_-_19772372 | 0.15 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr7_+_31879649 | 0.15 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_+_28124393 | 0.15 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr13_+_27314795 | 0.15 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr21_-_40676224 | 0.15 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr23_+_28770225 | 0.14 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr25_-_31953516 | 0.14 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr2_+_22531185 | 0.14 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr8_-_25338709 | 0.14 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr23_+_1730663 | 0.14 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr5_+_52067723 | 0.14 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr3_-_32934359 | 0.14 |
ENSDART00000124160
|
aoc2
|
amine oxidase, copper containing 2 |
| chr20_+_10723292 | 0.14 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr5_-_28038623 | 0.13 |
ENSDART00000160013
|
zgc:113436
|
zgc:113436 |
| chr19_-_3381422 | 0.13 |
ENSDART00000105146
|
edn1
|
endothelin 1 |
| chr19_-_10810006 | 0.13 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr15_-_6976851 | 0.13 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr15_+_14854666 | 0.13 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr11_+_30513656 | 0.13 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr2_+_32780138 | 0.13 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr10_-_16062565 | 0.12 |
ENSDART00000188728
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr22_+_38159823 | 0.12 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr15_-_14210382 | 0.12 |
ENSDART00000179599
|
CR925813.1
|
|
| chr16_+_52748263 | 0.12 |
ENSDART00000032275
|
atp6v1c1a
|
ATPase H+ transporting V1 subunit C1a |
| chr7_+_32722227 | 0.12 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr24_+_18299175 | 0.12 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr8_-_38403018 | 0.12 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr7_-_35409027 | 0.12 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr12_+_17754859 | 0.12 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr22_+_17120473 | 0.12 |
ENSDART00000170076
ENSDART00000090217 |
frem1b
|
Fras1 related extracellular matrix 1b |
| chr3_+_5504990 | 0.12 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr16_+_41570653 | 0.12 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr21_+_15824182 | 0.12 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr9_-_17417628 | 0.11 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr4_-_28958601 | 0.11 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr4_+_57099307 | 0.11 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr19_+_7495791 | 0.11 |
ENSDART00000010862
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chr17_+_42274825 | 0.11 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr2_-_3045861 | 0.11 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr5_-_64213253 | 0.11 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr4_+_14660769 | 0.11 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr7_-_10560964 | 0.11 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr4_+_912563 | 0.11 |
ENSDART00000103631
|
ripply2
|
ripply transcriptional repressor 2 |
| chr4_+_68476490 | 0.11 |
ENSDART00000168543
|
si:dkey-237g15.2
|
si:dkey-237g15.2 |
| chr19_+_14921000 | 0.10 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr1_-_38171648 | 0.10 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr9_+_14152211 | 0.10 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr6_-_55958705 | 0.10 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr10_+_43189325 | 0.10 |
ENSDART00000185584
|
vcanb
|
versican b |
| chr10_+_38708099 | 0.10 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr7_-_6470431 | 0.10 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr21_-_42055872 | 0.09 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr14_+_38845674 | 0.09 |
ENSDART00000129958
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr5_-_46152740 | 0.09 |
ENSDART00000123144
|
f2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr5_+_22459087 | 0.09 |
ENSDART00000134781
|
BX546499.1
|
|
| chr19_+_1370504 | 0.09 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr11_+_24716837 | 0.09 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr7_+_49664174 | 0.09 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr1_-_38170997 | 0.09 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr23_-_31512496 | 0.09 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr25_+_19095231 | 0.09 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr21_+_25198637 | 0.09 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr15_+_15779184 | 0.09 |
ENSDART00000156902
|
si:ch211-33e4.2
|
si:ch211-33e4.2 |
| chr7_+_3489195 | 0.09 |
ENSDART00000183876
|
si:ch211-285c6.3
|
si:ch211-285c6.3 |
| chr3_+_62205858 | 0.09 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr2_-_985417 | 0.08 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr20_+_43942278 | 0.08 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr13_+_15215633 | 0.08 |
ENSDART00000133041
|
pank2
|
pantothenate kinase 2 |
| chr5_-_38197080 | 0.08 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr13_+_22476742 | 0.08 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr8_+_17167876 | 0.08 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr2_+_45441563 | 0.08 |
ENSDART00000151882
|
si:ch211-66k16.27
|
si:ch211-66k16.27 |
| chr2_-_58257624 | 0.08 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr9_+_38420028 | 0.08 |
ENSDART00000135748
|
cyp27a1.2
|
cytochrome P450, family 27, subfamily A, polypeptide 1, gene 2 |
| chr19_-_29832876 | 0.08 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr3_+_13332870 | 0.08 |
ENSDART00000113878
|
CU138547.1
|
|
| chr15_-_14552101 | 0.08 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr25_+_7504314 | 0.08 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr5_+_21225017 | 0.07 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr10_+_9281991 | 0.07 |
ENSDART00000139156
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr14_-_38872536 | 0.07 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr6_+_39184236 | 0.07 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr4_-_62714083 | 0.07 |
ENSDART00000188299
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr2_-_57991638 | 0.07 |
ENSDART00000183937
|
si:dkeyp-68b7.12
|
si:dkeyp-68b7.12 |
| chr7_+_12931396 | 0.07 |
ENSDART00000184664
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr13_-_1130096 | 0.07 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr8_-_50888806 | 0.07 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr18_-_21859019 | 0.07 |
ENSDART00000100885
|
nrn1la
|
neuritin 1-like a |
| chr3_-_3939785 | 0.07 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr17_-_45734231 | 0.07 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr4_-_75707991 | 0.07 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr21_-_25618175 | 0.07 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr2_+_26656283 | 0.07 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr8_-_13210959 | 0.07 |
ENSDART00000142224
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr2_-_51794472 | 0.07 |
ENSDART00000186652
|
BX908782.3
|
|
| chr22_-_20733822 | 0.07 |
ENSDART00000133780
|
si:dkey-3k20.1
|
si:dkey-3k20.1 |
| chr4_-_73136420 | 0.07 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr6_+_18321627 | 0.07 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr9_-_9998087 | 0.06 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr9_+_38408991 | 0.06 |
ENSDART00000045588
|
cyp27a7
|
cytochrome P450, family 27, subfamily A, polypeptide 7 |
| chr23_-_26652273 | 0.06 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr4_+_66853199 | 0.06 |
ENSDART00000170223
|
si:dkey-29m1.2
|
si:dkey-29m1.2 |
| chr16_+_24626448 | 0.06 |
ENSDART00000153591
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr14_+_16813816 | 0.06 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr19_-_10648523 | 0.06 |
ENSDART00000059358
|
tekt2
|
tektin 2 (testicular) |
| chr19_+_9091673 | 0.06 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr1_+_49682312 | 0.06 |
ENSDART00000136747
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr4_-_76154252 | 0.06 |
ENSDART00000161269
|
BX957252.1
|
|
| chr5_-_16983336 | 0.06 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr10_+_40762200 | 0.06 |
ENSDART00000161000
ENSDART00000136185 |
taar19g
|
trace amine associated receptor 19g |
| chr11_-_43948885 | 0.06 |
ENSDART00000164522
|
CABZ01074203.1
|
|
| chr8_+_17168114 | 0.06 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr25_-_3469576 | 0.06 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr16_+_26017360 | 0.06 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr17_-_465285 | 0.06 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr1_+_12009673 | 0.06 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr24_-_1356668 | 0.06 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
| chr25_+_6186823 | 0.05 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042088 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.2 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.0 | 0.1 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.2 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.2 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 1.1 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.0 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.1 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |