Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
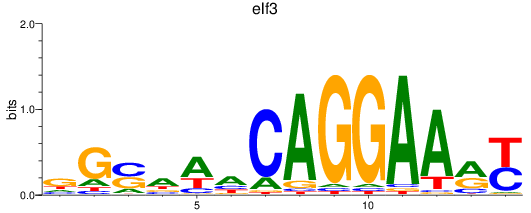
Results for elf3
Z-value: 0.52
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| elf3 | dr11_v1_chr22_+_661711_661711 | -0.51 | 1.6e-01 | Click! |
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_54290356 | 0.38 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr17_-_25330775 | 0.37 |
ENSDART00000154048
|
zpcx
|
zona pellucida protein C |
| chr16_-_15387459 | 0.35 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr15_-_37929135 | 0.34 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr23_+_32101361 | 0.32 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr12_+_30705769 | 0.30 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr23_+_32101202 | 0.29 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr23_+_26026383 | 0.28 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr17_+_24064014 | 0.28 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr14_-_28566238 | 0.27 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr5_-_29152457 | 0.26 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr12_+_30706158 | 0.26 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr5_+_40835601 | 0.25 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr19_-_20403845 | 0.25 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr16_+_29509133 | 0.24 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr10_+_38417512 | 0.24 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr13_-_33700461 | 0.24 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr2_+_31838442 | 0.24 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr5_-_54712159 | 0.24 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr7_+_35075847 | 0.23 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr2_-_24068848 | 0.23 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr1_-_24349759 | 0.23 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr5_-_24245218 | 0.22 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr19_-_19442983 | 0.21 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr4_-_20081621 | 0.20 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr19_+_15443353 | 0.19 |
ENSDART00000135923
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_+_41318412 | 0.19 |
ENSDART00000064614
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr14_-_28568107 | 0.19 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr21_+_3941758 | 0.19 |
ENSDART00000181345
|
setx
|
senataxin |
| chr15_+_14854666 | 0.19 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr3_-_15080226 | 0.19 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr13_+_41917606 | 0.18 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr6_-_55423220 | 0.18 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr13_-_51846224 | 0.18 |
ENSDART00000184663
|
LT631684.2
|
|
| chr14_+_22076596 | 0.17 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr19_+_15443063 | 0.17 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_-_33859950 | 0.17 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr14_+_41318604 | 0.17 |
ENSDART00000167042
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr23_+_32335871 | 0.17 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr5_+_37406358 | 0.17 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr2_+_3823813 | 0.17 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr7_+_16509201 | 0.17 |
ENSDART00000173777
|
zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr12_+_33395748 | 0.16 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr2_-_37462462 | 0.16 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr15_-_26570948 | 0.16 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr11_+_44226200 | 0.16 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr16_+_29514473 | 0.16 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr3_+_34159192 | 0.16 |
ENSDART00000151119
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr9_+_24125445 | 0.15 |
ENSDART00000090346
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr4_+_6735053 | 0.15 |
ENSDART00000150389
|
tmem168a
|
transmembrane protein 168a |
| chr5_-_29531948 | 0.15 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr4_-_7869731 | 0.15 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr5_-_57652168 | 0.15 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr6_-_44161262 | 0.15 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr4_-_59417501 | 0.15 |
ENSDART00000163357
|
BX321871.1
|
|
| chr4_-_17263210 | 0.15 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr19_+_43341115 | 0.15 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
| chr16_+_28727383 | 0.14 |
ENSDART00000149753
|
dcst2
|
DC-STAMP domain containing 2 |
| chr6_+_3717613 | 0.14 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr5_+_37087583 | 0.14 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr5_-_57311037 | 0.14 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr2_-_55779927 | 0.14 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr8_+_8947623 | 0.13 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr13_-_34858500 | 0.13 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr3_+_34234029 | 0.13 |
ENSDART00000044859
|
znf207a
|
zinc finger protein 207, a |
| chr22_+_1170294 | 0.13 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr17_-_21249651 | 0.13 |
ENSDART00000156580
ENSDART00000155221 |
hspa12a
|
heat shock protein 12A |
| chr1_-_34450622 | 0.13 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr10_-_1523253 | 0.13 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr5_-_39171302 | 0.13 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr15_-_8856391 | 0.13 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr17_-_11418513 | 0.13 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr19_+_43341424 | 0.13 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr4_+_68393422 | 0.13 |
ENSDART00000190956
|
CT737127.1
|
|
| chr5_-_69482891 | 0.12 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr24_+_6353394 | 0.12 |
ENSDART00000165118
|
CR352329.1
|
|
| chr16_-_40426837 | 0.12 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr14_-_30905288 | 0.12 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr21_+_5932140 | 0.12 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr10_+_29771256 | 0.12 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr15_+_21882419 | 0.12 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr2_+_16696052 | 0.12 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr6_-_19351495 | 0.12 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr20_+_13141408 | 0.12 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr20_-_33705044 | 0.12 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr22_-_17652112 | 0.12 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr12_+_28955766 | 0.12 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr4_-_4261673 | 0.12 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr13_-_21672131 | 0.12 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr9_+_34952203 | 0.12 |
ENSDART00000121828
ENSDART00000142347 |
tfdp1a
|
transcription factor Dp-1, a |
| chr22_-_17652914 | 0.12 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr20_+_25904199 | 0.12 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr18_-_40753583 | 0.12 |
ENSDART00000026767
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_+_431208 | 0.12 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr22_-_16758438 | 0.11 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr15_-_5799170 | 0.11 |
ENSDART00000142334
ENSDART00000171528 |
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr15_-_2734560 | 0.11 |
ENSDART00000153853
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr8_-_11202378 | 0.11 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr2_-_7246338 | 0.11 |
ENSDART00000186735
|
zgc:153115
|
zgc:153115 |
| chr20_-_34028967 | 0.11 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr19_-_1002959 | 0.11 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr1_+_10318089 | 0.11 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr6_-_15065376 | 0.11 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr5_-_25733745 | 0.11 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr5_-_44286778 | 0.11 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr4_-_17823424 | 0.11 |
ENSDART00000024822
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr22_+_2598998 | 0.11 |
ENSDART00000176665
|
CU570894.1
|
|
| chr20_-_33556983 | 0.11 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr13_+_1089942 | 0.11 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr3_-_42016693 | 0.11 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr13_+_47007075 | 0.11 |
ENSDART00000109247
ENSDART00000183205 ENSDART00000180924 ENSDART00000133146 |
anapc1
|
anaphase promoting complex subunit 1 |
| chr15_-_31265375 | 0.11 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr2_+_5406236 | 0.11 |
ENSDART00000154167
|
sft2d3
|
SFT2 domain containing 3 |
| chr4_+_51347000 | 0.11 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr5_-_37997774 | 0.10 |
ENSDART00000139616
ENSDART00000167694 |
si:dkey-111e8.1
|
si:dkey-111e8.1 |
| chr11_+_45343245 | 0.10 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr14_-_897874 | 0.10 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr21_+_6114305 | 0.10 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr23_-_10175898 | 0.10 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr1_+_36722122 | 0.10 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr12_-_33558727 | 0.10 |
ENSDART00000086087
|
mbtd1
|
mbt domain containing 1 |
| chr3_-_24205339 | 0.10 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr12_-_33558879 | 0.10 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr3_-_35865040 | 0.10 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr17_-_33416020 | 0.10 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr21_+_35328025 | 0.10 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr1_+_12301913 | 0.10 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr6_-_6254432 | 0.10 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr5_-_44286987 | 0.10 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr24_+_22485710 | 0.10 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr2_+_36721357 | 0.10 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr24_+_37449893 | 0.10 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr4_-_17353100 | 0.10 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr9_-_49860756 | 0.10 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr22_-_36934040 | 0.10 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr3_+_42999844 | 0.09 |
ENSDART00000183950
|
ints1
|
integrator complex subunit 1 |
| chr20_-_22798794 | 0.09 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr22_+_26600834 | 0.09 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr15_-_25349809 | 0.09 |
ENSDART00000152137
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr4_-_18840919 | 0.09 |
ENSDART00000015834
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr12_+_28910762 | 0.09 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr16_+_38201840 | 0.09 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr16_+_25171832 | 0.09 |
ENSDART00000156416
|
wu:fe05a04
|
wu:fe05a04 |
| chr6_-_28961660 | 0.09 |
ENSDART00000147285
|
tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr3_+_19665319 | 0.09 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr15_-_8856785 | 0.09 |
ENSDART00000192816
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr4_-_18841071 | 0.09 |
ENSDART00000140722
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr2_-_2096055 | 0.09 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_+_41748693 | 0.09 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr9_+_12948511 | 0.09 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr10_-_41450367 | 0.09 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr13_-_35908275 | 0.09 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr10_+_40324395 | 0.09 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr3_+_19245804 | 0.09 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr3_-_40965328 | 0.09 |
ENSDART00000102393
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr11_+_44236183 | 0.09 |
ENSDART00000193470
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr20_-_2619316 | 0.09 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr5_-_4297459 | 0.09 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr7_+_26545502 | 0.09 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr21_+_25801345 | 0.08 |
ENSDART00000035062
|
nf2b
|
neurofibromin 2b (merlin) |
| chr11_-_18601955 | 0.08 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr2_-_7246848 | 0.08 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr5_+_61944453 | 0.08 |
ENSDART00000134344
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr19_+_9111550 | 0.08 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr9_-_23147026 | 0.08 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr20_+_39250673 | 0.08 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr17_-_5610514 | 0.08 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr20_+_32481348 | 0.08 |
ENSDART00000185018
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr18_+_45862414 | 0.08 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr20_+_27087539 | 0.08 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr21_-_41065369 | 0.08 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr20_+_34501152 | 0.08 |
ENSDART00000143765
|
gorab
|
golgin, rab6-interacting |
| chr8_+_8712446 | 0.08 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr9_+_6079528 | 0.08 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr20_-_43572763 | 0.08 |
ENSDART00000153251
|
pimr125
|
Pim proto-oncogene, serine/threonine kinase, related 125 |
| chr8_-_13315304 | 0.08 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr2_+_36109002 | 0.08 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr5_-_23464970 | 0.08 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr5_-_13206878 | 0.08 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr22_-_23781083 | 0.08 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr23_-_4705110 | 0.08 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr22_+_3238474 | 0.08 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr20_-_1191910 | 0.08 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr3_-_40965596 | 0.08 |
ENSDART00000137188
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr13_+_33268657 | 0.08 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr17_+_19626479 | 0.08 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr7_+_5904020 | 0.08 |
ENSDART00000172847
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr7_+_73295890 | 0.08 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr2_-_27651674 | 0.07 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr15_-_2754056 | 0.07 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr24_+_26140855 | 0.07 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr6_-_19665114 | 0.07 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr14_-_237130 | 0.07 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr15_+_1534644 | 0.07 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr1_-_44484 | 0.07 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr10_+_17713022 | 0.07 |
ENSDART00000170453
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr1_-_46718353 | 0.07 |
ENSDART00000074514
|
spryd7a
|
SPRY domain containing 7a |
| chr14_-_26436951 | 0.07 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr22_+_2300484 | 0.07 |
ENSDART00000106503
|
znf1180
|
zinc finger protein 1180 |
| chr7_+_31051603 | 0.07 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.0 | 0.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.0 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.0 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |