Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
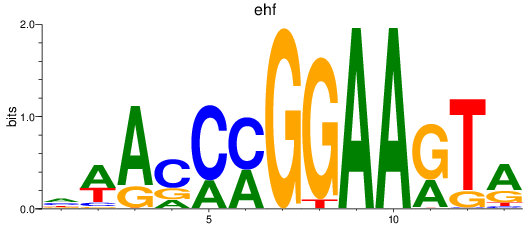
Results for ehf
Z-value: 0.52
Transcription factors associated with ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ehf
|
ENSDARG00000052115 | ets homologous factor |
|
ehf
|
ENSDARG00000112589 | ets homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ehf | dr11_v1_chr25_+_10410620_10410620 | -0.76 | 1.8e-02 | Click! |
Activity profile of ehf motif
Sorted Z-values of ehf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_9997948 | 0.69 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_10014512 | 0.65 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr21_-_43649959 | 0.61 |
ENSDART00000142733
|
si:dkey-229d11.5
|
si:dkey-229d11.5 |
| chr17_-_25331439 | 0.58 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr16_-_46393154 | 0.54 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr15_-_35126332 | 0.54 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr1_-_8553165 | 0.48 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr21_+_3941758 | 0.47 |
ENSDART00000181345
|
setx
|
senataxin |
| chr10_-_1961576 | 0.47 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr21_-_43665537 | 0.46 |
ENSDART00000157610
|
si:dkey-229d11.3
|
si:dkey-229d11.3 |
| chr21_-_43666420 | 0.44 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr20_+_33987465 | 0.43 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr5_+_40835601 | 0.42 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr8_+_41038141 | 0.40 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr5_-_54712159 | 0.40 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr12_-_9468618 | 0.38 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr8_+_8643901 | 0.38 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr20_+_54027778 | 0.37 |
ENSDART00000060458
|
si:dkey-241l7.3
|
si:dkey-241l7.3 |
| chr13_-_25408387 | 0.36 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr14_-_897874 | 0.35 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr24_+_31459715 | 0.35 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr23_+_17522867 | 0.35 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr22_-_17653143 | 0.35 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr10_-_26202766 | 0.35 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr7_-_58178807 | 0.34 |
ENSDART00000188531
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr8_+_41037541 | 0.34 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr4_-_5826320 | 0.33 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr3_+_34120191 | 0.32 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr17_+_24809221 | 0.32 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr10_+_40324395 | 0.32 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr22_-_17652914 | 0.31 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr3_+_36617024 | 0.31 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr1_-_53468160 | 0.31 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr13_+_39282477 | 0.31 |
ENSDART00000132198
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr14_+_30272891 | 0.31 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr13_-_22961605 | 0.31 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr21_-_9769500 | 0.30 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_25084385 | 0.30 |
ENSDART00000134526
ENSDART00000111863 |
paxx
|
PAXX, non-homologous end joining factor |
| chr25_+_5015019 | 0.30 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr17_-_31212420 | 0.29 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr4_-_17263210 | 0.29 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr17_-_9962578 | 0.28 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr20_+_51813432 | 0.28 |
ENSDART00000127444
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr7_-_58178980 | 0.28 |
ENSDART00000073635
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr8_+_622640 | 0.28 |
ENSDART00000051774
|
spinb
|
spindlin b |
| chr6_+_13045885 | 0.28 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr18_+_45862414 | 0.28 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr15_+_34933552 | 0.27 |
ENSDART00000155368
|
zgc:66024
|
zgc:66024 |
| chr12_-_33359654 | 0.27 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_+_50150834 | 0.27 |
ENSDART00000056074
|
entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr8_+_48965767 | 0.27 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr17_+_10578823 | 0.27 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr3_+_19665319 | 0.26 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr22_-_16758438 | 0.26 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr20_+_46572550 | 0.26 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr20_+_23947004 | 0.26 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr13_+_25549425 | 0.26 |
ENSDART00000087553
ENSDART00000169199 |
sec23ip
|
SEC23 interacting protein |
| chr5_+_19933356 | 0.25 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr1_+_19649545 | 0.25 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr5_-_16475682 | 0.25 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr7_+_13684012 | 0.25 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr21_+_20386865 | 0.25 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr8_+_41035230 | 0.25 |
ENSDART00000141349
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr21_-_22117085 | 0.25 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr8_+_48966165 | 0.25 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr3_-_40051425 | 0.24 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr6_+_12527725 | 0.24 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr8_-_11131695 | 0.24 |
ENSDART00000055742
|
nras
|
NRAS proto-oncogene, GTPase |
| chr11_-_25853212 | 0.24 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr20_+_13141408 | 0.24 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr23_-_26488622 | 0.23 |
ENSDART00000168052
ENSDART00000169088 |
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr16_-_25233515 | 0.23 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr24_-_1010467 | 0.23 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr16_+_51462620 | 0.22 |
ENSDART00000169443
|
SLC9A1
|
solute carrier family 9 member A1 |
| chr3_+_36616713 | 0.22 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr2_+_30182431 | 0.22 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr17_-_20218167 | 0.22 |
ENSDART00000154479
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr25_-_25575241 | 0.22 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr25_-_25575717 | 0.22 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr20_+_34501152 | 0.22 |
ENSDART00000143765
|
gorab
|
golgin, rab6-interacting |
| chr11_+_2596667 | 0.21 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr4_-_17257435 | 0.21 |
ENSDART00000131973
|
lrmp
|
lymphoid-restricted membrane protein |
| chr10_-_41302841 | 0.21 |
ENSDART00000020297
ENSDART00000160174 ENSDART00000183850 ENSDART00000169493 |
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr14_-_28568107 | 0.21 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr17_-_31611692 | 0.21 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr14_-_5407555 | 0.21 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr16_-_29690188 | 0.21 |
ENSDART00000132351
ENSDART00000004284 |
scnm1
|
sodium channel modifier 1 |
| chr9_-_15789526 | 0.20 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr4_+_17671614 | 0.20 |
ENSDART00000121714
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr19_+_11432330 | 0.20 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr7_-_6754012 | 0.20 |
ENSDART00000113658
|
zgc:55262
|
zgc:55262 |
| chr6_-_44161262 | 0.20 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr21_-_19919918 | 0.19 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_+_35424539 | 0.19 |
ENSDART00000171809
ENSDART00000162185 |
sytl4
|
synaptotagmin-like 4 |
| chr14_-_43616572 | 0.19 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr2_+_37207461 | 0.19 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr19_+_43523690 | 0.19 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr17_-_20218357 | 0.19 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr5_-_35264517 | 0.19 |
ENSDART00000114981
|
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr22_+_12161738 | 0.19 |
ENSDART00000168857
|
ccnt2b
|
cyclin T2b |
| chr9_+_29520696 | 0.19 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr18_+_22217489 | 0.19 |
ENSDART00000165464
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr25_-_3745393 | 0.19 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr25_+_3549841 | 0.19 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr14_-_20891745 | 0.19 |
ENSDART00000106205
|
ids
|
iduronate 2-sulfatase |
| chr24_-_37326236 | 0.19 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr3_+_27664864 | 0.19 |
ENSDART00000126533
ENSDART00000180848 |
clcn7
|
chloride channel 7 |
| chr15_-_34933560 | 0.19 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr15_-_34878388 | 0.19 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr20_-_3238110 | 0.19 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr3_-_58455289 | 0.18 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr15_+_17100697 | 0.18 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_+_3823813 | 0.18 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr13_-_24396003 | 0.18 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr3_-_35865040 | 0.18 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr8_-_26709959 | 0.18 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr12_+_47794089 | 0.18 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr16_-_42770064 | 0.18 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr4_+_17671164 | 0.18 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr15_-_43284021 | 0.18 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr18_-_39186262 | 0.18 |
ENSDART00000029549
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr7_+_19850889 | 0.18 |
ENSDART00000100782
|
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr3_-_21061931 | 0.18 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr17_-_24879003 | 0.18 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr20_-_48898371 | 0.18 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr8_+_17529151 | 0.18 |
ENSDART00000147760
|
si:ch73-70k4.1
|
si:ch73-70k4.1 |
| chr8_-_20862443 | 0.18 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr13_-_24396199 | 0.18 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr7_-_48391571 | 0.18 |
ENSDART00000180034
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr14_+_14806692 | 0.18 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr11_+_25157374 | 0.18 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr1_-_34685329 | 0.18 |
ENSDART00000125944
ENSDART00000008277 |
pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr20_-_3310017 | 0.18 |
ENSDART00000099315
|
cebpz
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr14_-_32631013 | 0.18 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr1_+_12301913 | 0.18 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr15_-_28802140 | 0.17 |
ENSDART00000156120
ENSDART00000156708 ENSDART00000122536 ENSDART00000155008 ENSDART00000153825 |
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr13_+_9559461 | 0.17 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr20_-_54522660 | 0.17 |
ENSDART00000059872
|
ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr10_+_36441124 | 0.17 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr3_+_46479913 | 0.17 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr7_+_36472789 | 0.17 |
ENSDART00000136545
|
aktip
|
akt interacting protein |
| chr13_-_43637371 | 0.17 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr7_+_19851422 | 0.17 |
ENSDART00000142970
ENSDART00000190027 |
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr7_+_24573721 | 0.17 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr6_-_40713183 | 0.17 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr16_+_3982590 | 0.17 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr3_-_402714 | 0.17 |
ENSDART00000134062
ENSDART00000105659 |
mhc1zja
|
major histocompatibility complex class I ZJA |
| chr17_+_37253706 | 0.17 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr16_-_40426837 | 0.17 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr25_+_29688671 | 0.17 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr7_-_59165640 | 0.17 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr5_-_67629263 | 0.17 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr8_-_25605537 | 0.17 |
ENSDART00000005906
|
stk38a
|
serine/threonine kinase 38a |
| chr3_+_27665160 | 0.17 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr7_-_30605089 | 0.16 |
ENSDART00000173775
ENSDART00000173789 ENSDART00000166046 ENSDART00000111626 |
rnf111
|
ring finger protein 111 |
| chr3_-_30186296 | 0.16 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr5_+_50953240 | 0.16 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr9_-_27719998 | 0.16 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr10_+_29771256 | 0.16 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr1_+_27977297 | 0.16 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr1_+_46405294 | 0.16 |
ENSDART00000143228
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr21_+_6114305 | 0.16 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr7_-_69185124 | 0.16 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr19_-_15420678 | 0.16 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr10_-_7636340 | 0.16 |
ENSDART00000166234
|
ubxn8
|
UBX domain protein 8 |
| chr9_-_20853439 | 0.16 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr15_-_45510977 | 0.16 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr1_-_41374117 | 0.16 |
ENSDART00000074777
|
htt
|
huntingtin |
| chr2_+_38055529 | 0.16 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr19_+_37135700 | 0.16 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr14_-_5407118 | 0.16 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr24_-_36187094 | 0.16 |
ENSDART00000111891
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr11_-_21404044 | 0.15 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr7_-_13906409 | 0.15 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr8_-_47329755 | 0.15 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr7_+_39006837 | 0.15 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_+_47618221 | 0.15 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr5_+_19867794 | 0.15 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr8_-_621957 | 0.15 |
ENSDART00000050352
|
si:ch211-87m7.2
|
si:ch211-87m7.2 |
| chr15_-_28904371 | 0.15 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_19379084 | 0.15 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr3_-_5067585 | 0.15 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr22_+_3184500 | 0.15 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr6_+_40554551 | 0.15 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr22_-_38951128 | 0.15 |
ENSDART00000085685
ENSDART00000104422 |
rbbp9
|
retinoblastoma binding protein 9 |
| chr22_+_10163901 | 0.15 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr7_+_69528850 | 0.15 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr11_+_13176568 | 0.15 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr2_+_5406236 | 0.15 |
ENSDART00000154167
|
sft2d3
|
SFT2 domain containing 3 |
| chr5_+_61944453 | 0.15 |
ENSDART00000134344
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr1_-_55044256 | 0.15 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr9_+_28232522 | 0.15 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr5_-_12743196 | 0.15 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr3_-_57630791 | 0.15 |
ENSDART00000129598
|
usp36
|
ubiquitin specific peptidase 36 |
| chr25_+_19238175 | 0.15 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr14_-_38946808 | 0.15 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr5_+_22459087 | 0.15 |
ENSDART00000134781
|
BX546499.1
|
|
| chr22_+_2058394 | 0.15 |
ENSDART00000177038
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr2_+_2563192 | 0.15 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr23_+_4709607 | 0.15 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ehf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 1.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.5 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.2 | 0.5 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.1 | 0.3 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.1 | 0.3 | GO:0009193 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.3 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:0033152 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) immunoglobulin V(D)J recombination(GO:0033152) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.2 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.1 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.0 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.2 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 1.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.2 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.3 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.2 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.0 | 0.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.0 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |