Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
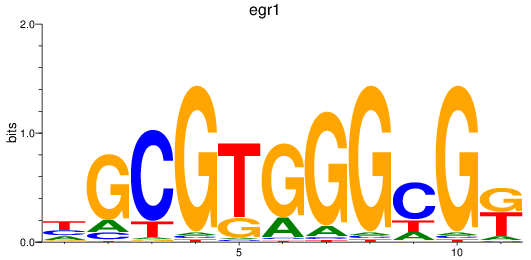
Results for egr1
Z-value: 0.98
Transcription factors associated with egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr1
|
ENSDARG00000037421 | early growth response 1 |
|
egr1
|
ENSDARG00000115563 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr1 | dr11_v1_chr14_-_21336385_21336385 | -0.67 | 4.7e-02 | Click! |
Activity profile of egr1 motif
Sorted Z-values of egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_15155684 | 2.88 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr10_-_34002185 | 2.77 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr19_-_6193448 | 2.38 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr15_-_17099560 | 2.14 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr3_-_49504023 | 2.09 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr19_-_6193067 | 2.04 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr17_+_46387086 | 1.68 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr10_-_32851847 | 1.64 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr9_+_45227028 | 1.63 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr22_+_34784075 | 1.55 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr17_-_2584423 | 1.35 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_2115040 | 1.31 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr21_+_19368720 | 1.30 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr18_+_15106518 | 1.29 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr7_-_20611039 | 1.25 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr23_-_22072460 | 1.24 |
ENSDART00000132343
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr9_-_54001502 | 1.21 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr24_+_26379441 | 1.20 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr4_-_277081 | 1.20 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr2_-_24603325 | 1.17 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr20_+_27712714 | 1.16 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr25_+_34889061 | 1.15 |
ENSDART00000136226
|
spire2
|
spire-type actin nucleation factor 2 |
| chr23_-_22072716 | 1.12 |
ENSDART00000054362
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr2_+_35880600 | 1.12 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr11_+_2855430 | 1.10 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr1_-_17693273 | 1.09 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr7_+_46019780 | 1.09 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr16_+_13965923 | 1.09 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr15_-_14038227 | 1.07 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr2_+_50062165 | 1.06 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr5_+_37720777 | 1.05 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr1_-_8917902 | 1.05 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr9_-_41401564 | 1.05 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr13_-_21739142 | 0.98 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr2_+_38924975 | 0.98 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr18_+_3169579 | 0.96 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr12_+_47448558 | 0.95 |
ENSDART00000185689
ENSDART00000105328 |
fmn2b
|
formin 2b |
| chr23_-_31060350 | 0.94 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr23_+_35426404 | 0.93 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr8_+_28724692 | 0.92 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr7_+_20563305 | 0.89 |
ENSDART00000169661
|
si:dkey-19b23.10
|
si:dkey-19b23.10 |
| chr12_-_48943467 | 0.88 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr21_+_3941758 | 0.88 |
ENSDART00000181345
|
setx
|
senataxin |
| chr25_-_36370292 | 0.87 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr8_+_2642384 | 0.86 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr21_-_43398457 | 0.86 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr17_+_34206167 | 0.84 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr23_+_45822935 | 0.83 |
ENSDART00000161892
|
vdra
|
vitamin D receptor a |
| chr1_+_58977262 | 0.83 |
ENSDART00000168832
|
eri2
|
ERI1 exoribonuclease family member 2 |
| chr20_+_39250673 | 0.83 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr17_-_25831569 | 0.83 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr16_-_13965518 | 0.81 |
ENSDART00000138304
|
si:dkey-85k15.4
|
si:dkey-85k15.4 |
| chr18_-_50845804 | 0.80 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr25_+_34888886 | 0.79 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr10_-_14920989 | 0.79 |
ENSDART00000184617
|
smad2
|
SMAD family member 2 |
| chr11_-_37880492 | 0.78 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr21_-_41369539 | 0.78 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr14_-_4044545 | 0.78 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr5_-_67241633 | 0.77 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr14_+_17397534 | 0.76 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr22_-_16275236 | 0.75 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr3_+_16722014 | 0.74 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr2_+_23062085 | 0.74 |
ENSDART00000153745
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr24_+_19542323 | 0.73 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr2_+_38271392 | 0.73 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr19_-_25427255 | 0.72 |
ENSDART00000036854
|
glcci1a
|
glucocorticoid induced 1a |
| chr21_-_43398122 | 0.72 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr6_-_7776612 | 0.72 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr7_+_9326234 | 0.72 |
ENSDART00000104536
|
chsy1
|
chondroitin sulfate synthase 1 |
| chr16_+_19029297 | 0.71 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr5_+_66170479 | 0.70 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr13_-_49666615 | 0.69 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr20_-_22778394 | 0.68 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr13_+_50778187 | 0.67 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr17_+_584369 | 0.64 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr12_-_37449396 | 0.64 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr11_+_1565806 | 0.64 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr9_+_51784922 | 0.63 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr2_+_105748 | 0.63 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr20_+_27087539 | 0.62 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr1_-_29761086 | 0.62 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr7_-_24995631 | 0.61 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr20_+_28803642 | 0.61 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr11_-_2594045 | 0.60 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr22_+_10713713 | 0.60 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr3_+_51684963 | 0.59 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr11_+_45461419 | 0.59 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr7_+_31051603 | 0.59 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr21_-_41369370 | 0.58 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr25_+_29688671 | 0.58 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr13_-_36761379 | 0.57 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr20_-_46808870 | 0.56 |
ENSDART00000190911
|
esrrgb
|
estrogen-related receptor gamma b |
| chr18_+_18612388 | 0.55 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr12_+_48511605 | 0.55 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr3_+_1211242 | 0.54 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr23_+_44665230 | 0.52 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr22_-_193234 | 0.52 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr7_+_28724919 | 0.52 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr23_-_16980213 | 0.52 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr9_-_105135 | 0.51 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr17_+_15983247 | 0.50 |
ENSDART00000154275
|
clmn
|
calmin |
| chr16_-_26132122 | 0.49 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr7_-_46019756 | 0.49 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr23_-_31645760 | 0.48 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr25_-_32888115 | 0.47 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr3_-_32180796 | 0.47 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr23_-_32100106 | 0.46 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr22_-_3564563 | 0.46 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr13_-_479129 | 0.46 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr7_+_31051213 | 0.45 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr4_-_5018705 | 0.45 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr20_+_32523576 | 0.44 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr12_+_41510492 | 0.44 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr12_+_34273240 | 0.44 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr3_-_42016693 | 0.44 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr25_+_36315127 | 0.42 |
ENSDART00000191984
|
zgc:165555
|
zgc:165555 |
| chr24_-_42072886 | 0.42 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr3_+_1107102 | 0.42 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr20_+_28803977 | 0.42 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_+_77948517 | 0.40 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_+_2271559 | 0.40 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr7_-_52498175 | 0.38 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr20_+_46699021 | 0.38 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr11_-_42418374 | 0.37 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr2_+_23701613 | 0.36 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr21_+_27513859 | 0.36 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr21_+_103194 | 0.36 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr5_+_72377851 | 0.36 |
ENSDART00000160479
|
lman2la
|
lectin, mannose-binding 2-like a |
| chr1_+_49435017 | 0.36 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr3_-_5228137 | 0.36 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr7_+_5968225 | 0.35 |
ENSDART00000083397
|
zgc:165555
|
zgc:165555 |
| chr2_+_20793982 | 0.35 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr1_+_59146298 | 0.35 |
ENSDART00000191885
ENSDART00000152747 |
gpr108
|
G protein-coupled receptor 108 |
| chr19_+_935565 | 0.34 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr1_-_10473630 | 0.33 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr4_-_5019113 | 0.33 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr20_+_38458084 | 0.33 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr10_+_11282883 | 0.32 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr1_+_12295142 | 0.32 |
ENSDART00000158595
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr11_+_45436703 | 0.32 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr17_+_15983557 | 0.32 |
ENSDART00000190806
|
clmn
|
calmin |
| chr25_-_6389713 | 0.31 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr23_-_270847 | 0.31 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr10_+_8554929 | 0.31 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr14_-_7245971 | 0.30 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr6_+_42338309 | 0.30 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr7_-_66126628 | 0.30 |
ENSDART00000184492
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr19_+_2619444 | 0.29 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr10_-_35030673 | 0.29 |
ENSDART00000135338
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr1_+_176583 | 0.29 |
ENSDART00000168760
ENSDART00000160425 |
LAMP1
|
lysosomal associated membrane protein 1 |
| chr12_-_850457 | 0.29 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr5_-_2689753 | 0.28 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr2_-_44344321 | 0.28 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr15_+_17406920 | 0.27 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr7_+_41340520 | 0.26 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr25_-_36310561 | 0.26 |
ENSDART00000181152
|
zgc:165555
|
zgc:165555 |
| chr14_+_164556 | 0.25 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr21_+_26620867 | 0.25 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr19_-_81477 | 0.25 |
ENSDART00000159815
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr21_-_24865217 | 0.25 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr5_-_21524251 | 0.24 |
ENSDART00000176793
ENSDART00000040184 |
tenm1
|
teneurin transmembrane protein 1 |
| chr17_+_8323348 | 0.24 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr2_+_16710889 | 0.23 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr6_-_25181012 | 0.23 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr2_-_54639964 | 0.23 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr8_+_20438884 | 0.22 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr11_+_25296366 | 0.22 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr21_-_33126697 | 0.22 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr14_+_48960078 | 0.21 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr6_+_14301214 | 0.21 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr7_-_26603743 | 0.20 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr19_+_2602903 | 0.19 |
ENSDART00000033132
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_-_52505279 | 0.19 |
ENSDART00000052907
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr6_-_13498745 | 0.19 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr2_-_50272077 | 0.19 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr17_+_20639180 | 0.19 |
ENSDART00000063624
|
npy4r
|
neuropeptide Y receptor Y4 |
| chr22_+_1751640 | 0.19 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr25_-_13050959 | 0.19 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr7_-_12596727 | 0.19 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr11_+_45461853 | 0.19 |
ENSDART00000173409
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr11_-_31146808 | 0.18 |
ENSDART00000172781
|
si:cabz01069582.1
|
si:cabz01069582.1 |
| chr1_-_7917062 | 0.18 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr24_+_32472155 | 0.18 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr17_-_14726824 | 0.18 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr22_+_15979430 | 0.18 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr2_+_59046036 | 0.17 |
ENSDART00000158860
|
stk11
|
serine/threonine kinase 11 |
| chr8_-_15292197 | 0.17 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr6_-_57408576 | 0.17 |
ENSDART00000163930
|
znfx1
|
zinc finger, NFX1-type containing 1 |
| chr25_-_7999756 | 0.17 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr20_-_26624042 | 0.17 |
ENSDART00000077962
|
exoc2
|
exocyst complex component 2 |
| chr22_-_6499055 | 0.17 |
ENSDART00000142235
ENSDART00000183588 |
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr14_-_15155384 | 0.16 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr17_+_53435279 | 0.16 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr19_+_7154500 | 0.15 |
ENSDART00000035967
ENSDART00000160894 |
brd2a
|
bromodomain containing 2a |
| chr19_-_3303995 | 0.14 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr22_-_10605045 | 0.14 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr6_+_34511886 | 0.14 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr5_+_65492183 | 0.14 |
ENSDART00000162804
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr16_-_8520474 | 0.13 |
ENSDART00000137365
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr6_+_13206516 | 0.13 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.6 | 1.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.1 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 2.1 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 0.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 0.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 1.7 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 1.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.7 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 1.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.8 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.7 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 1.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.7 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.8 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 2.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0044857 | membrane raft assembly(GO:0001765) membrane raft organization(GO:0031579) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 1.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.8 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.3 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 7.3 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0060349 | bone morphogenesis(GO:0060349) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 1.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 0.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.7 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 1.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 0.9 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 0.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.6 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 0.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 2.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 4.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 3.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |