Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
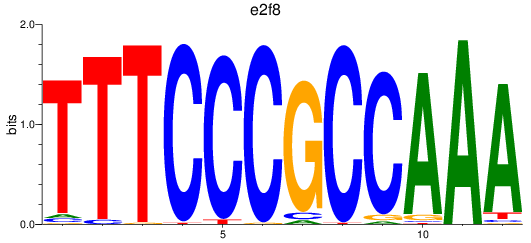
Results for e2f8
Z-value: 1.28
Transcription factors associated with e2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f8
|
ENSDARG00000057323 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f8 | dr11_v1_chr7_-_16596938_16596938 | 0.93 | 2.5e-04 | Click! |
Activity profile of e2f8 motif
Sorted Z-values of e2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_33220342 | 2.46 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr7_-_55648336 | 2.08 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr7_+_24814866 | 1.87 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr8_-_44238526 | 1.50 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr13_+_31402067 | 1.44 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr23_-_29357764 | 1.42 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr1_-_18811517 | 1.38 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr10_+_6884123 | 1.35 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr13_+_46941930 | 1.22 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr7_+_41812636 | 1.21 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr8_+_41038141 | 1.20 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr1_-_24558838 | 1.17 |
ENSDART00000102522
|
gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr6_+_21005725 | 1.13 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr7_+_41812817 | 1.12 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr7_-_39360325 | 1.12 |
ENSDART00000098033
ENSDART00000173695 ENSDART00000173466 ENSDART00000173734 |
ambra1a
|
autophagy/beclin-1 regulator 1a |
| chr16_-_9453591 | 1.11 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr9_+_34380299 | 1.09 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr4_-_7869731 | 1.09 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr1_-_34450622 | 1.07 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr7_-_6362460 | 1.02 |
ENSDART00000129239
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr19_+_40069524 | 1.02 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr19_+_42660158 | 1.00 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr7_+_41812190 | 1.00 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr14_-_28566238 | 0.98 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr16_+_33953644 | 0.98 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr11_-_26576754 | 0.94 |
ENSDART00000191733
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr7_-_73854476 | 0.92 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr7_-_73845736 | 0.92 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr22_-_4439311 | 0.91 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr20_+_13141408 | 0.87 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr22_-_38274188 | 0.86 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr21_-_11655010 | 0.86 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr7_+_5956937 | 0.85 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr13_-_14929236 | 0.84 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr14_+_49220026 | 0.84 |
ENSDART00000063643
ENSDART00000128744 |
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr3_-_35865040 | 0.83 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr11_-_11792766 | 0.83 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr23_-_16981899 | 0.82 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr5_-_30382925 | 0.81 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr16_+_31942527 | 0.80 |
ENSDART00000188334
ENSDART00000188643 ENSDART00000058496 |
phc1
|
polyhomeotic homolog 1 |
| chr19_-_2582858 | 0.79 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr14_-_32631519 | 0.79 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr9_+_34232503 | 0.78 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr21_-_41818359 | 0.77 |
ENSDART00000046076
|
CU929230.1
|
|
| chr16_+_53278406 | 0.77 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr1_-_45614318 | 0.76 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_-_18531760 | 0.75 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr20_+_53368611 | 0.74 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr22_+_26703026 | 0.73 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr7_-_804515 | 0.73 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr23_-_33738945 | 0.72 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr11_+_24804556 | 0.71 |
ENSDART00000082761
ENSDART00000186283 |
adipor1a
|
adiponectin receptor 1a |
| chr6_+_38896158 | 0.70 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr3_-_53092509 | 0.70 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr24_-_13349802 | 0.70 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr16_-_39195318 | 0.69 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr25_-_35110695 | 0.69 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr13_+_48358467 | 0.69 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr5_+_22459087 | 0.69 |
ENSDART00000134781
|
BX546499.1
|
|
| chr4_-_2380173 | 0.68 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr14_-_31465905 | 0.66 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr13_+_2894536 | 0.64 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr16_+_25116827 | 0.64 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr23_+_39413163 | 0.64 |
ENSDART00000184254
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr8_+_8196087 | 0.64 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr22_+_18349794 | 0.63 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr4_-_73488406 | 0.62 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr25_+_35133404 | 0.60 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr5_+_13308139 | 0.60 |
ENSDART00000182189
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr9_-_2892045 | 0.60 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr7_+_71535045 | 0.59 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr6_-_33916756 | 0.59 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr24_-_30843250 | 0.59 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr21_-_11970199 | 0.59 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_+_41421998 | 0.59 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr9_-_48407408 | 0.59 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr6_-_8466717 | 0.58 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr3_+_14571813 | 0.58 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr20_-_38827623 | 0.58 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr25_-_26753196 | 0.57 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr2_+_9755422 | 0.56 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr13_-_45942000 | 0.56 |
ENSDART00000158534
ENSDART00000171822 ENSDART00000166442 |
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr2_-_53500424 | 0.56 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr15_-_4596623 | 0.56 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr24_-_13349464 | 0.55 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr11_-_18017918 | 0.55 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr13_-_18011168 | 0.55 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr22_+_8753092 | 0.55 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr14_-_33351103 | 0.55 |
ENSDART00000010019
|
upf3b
|
UPF3B, regulator of nonsense mediated mRNA decay |
| chr7_+_26545502 | 0.54 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr20_-_38525467 | 0.54 |
ENSDART00000061417
|
si:ch211-245h14.1
|
si:ch211-245h14.1 |
| chr25_-_35102781 | 0.54 |
ENSDART00000180881
ENSDART00000153747 |
si:dkey-108k21.24
|
si:dkey-108k21.24 |
| chr7_-_6355459 | 0.54 |
ENSDART00000172898
|
CU457819.1
|
|
| chr5_-_20678300 | 0.53 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr7_-_6345507 | 0.53 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr3_+_52545400 | 0.53 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr22_+_8753365 | 0.52 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr11_-_18107447 | 0.52 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr17_+_11372531 | 0.51 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr11_-_18017287 | 0.51 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr7_+_26545911 | 0.51 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr23_+_10805188 | 0.51 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr25_-_35140746 | 0.49 |
ENSDART00000129969
|
si:ch211-113a14.19
|
si:ch211-113a14.19 |
| chr6_+_21062416 | 0.49 |
ENSDART00000141643
|
si:dkey-91f15.1
|
si:dkey-91f15.1 |
| chr11_+_45092866 | 0.48 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr13_+_48359573 | 0.47 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr25_-_35145920 | 0.46 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr12_+_17933775 | 0.46 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr9_-_2892250 | 0.46 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr2_+_37424261 | 0.45 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr3_-_26806032 | 0.44 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_-_49964810 | 0.44 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr1_+_59142783 | 0.43 |
ENSDART00000152669
|
gpr108
|
G protein-coupled receptor 108 |
| chr5_-_64454459 | 0.43 |
ENSDART00000172321
ENSDART00000168030 |
brd3b
|
bromodomain containing 3b |
| chr6_+_50451337 | 0.43 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr2_-_6104035 | 0.42 |
ENSDART00000053867
|
zte38
|
zebrafish testis-expressed 38 |
| chr16_-_12723738 | 0.42 |
ENSDART00000080414
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr11_+_3005536 | 0.42 |
ENSDART00000174539
|
cpne5b
|
copine Vb |
| chr16_-_43011470 | 0.42 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr13_-_18069421 | 0.42 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr7_-_6415991 | 0.41 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr25_-_35109536 | 0.41 |
ENSDART00000185229
|
CU302436.5
|
|
| chr12_+_21299338 | 0.41 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr3_-_14571514 | 0.39 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr10_-_37487612 | 0.39 |
ENSDART00000167964
ENSDART00000171326 |
si:dkey-65j4.2
|
si:dkey-65j4.2 |
| chr11_+_3006124 | 0.38 |
ENSDART00000126071
|
cpne5b
|
copine Vb |
| chr24_-_26945390 | 0.36 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr3_+_43086548 | 0.36 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr22_-_22301672 | 0.36 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr6_+_18531932 | 0.35 |
ENSDART00000165271
|
suz12b
|
SUZ12 polycomb repressive complex 2b subunit |
| chr19_-_32042105 | 0.35 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr3_+_52545014 | 0.35 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr5_-_28016805 | 0.35 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr20_+_18703454 | 0.35 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr13_+_26703922 | 0.34 |
ENSDART00000020946
|
fancl
|
Fanconi anemia, complementation group L |
| chr16_-_22775480 | 0.34 |
ENSDART00000141778
ENSDART00000145585 ENSDART00000125963 ENSDART00000127570 |
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr10_+_573667 | 0.34 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr18_+_45708744 | 0.34 |
ENSDART00000077341
|
depdc7
|
DEP domain containing 7 |
| chr15_-_37104165 | 0.33 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr4_+_13953537 | 0.32 |
ENSDART00000133596
|
pphln1
|
periphilin 1 |
| chr13_-_1423008 | 0.31 |
ENSDART00000110828
|
znf451
|
zinc finger protein 451 |
| chr6_-_28345002 | 0.31 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr1_-_46505310 | 0.30 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr19_-_30359059 | 0.29 |
ENSDART00000103471
|
khdrbs1b
|
KH domain containing, RNA binding, signal transduction associated 1b |
| chr20_+_41859277 | 0.29 |
ENSDART00000061141
|
cep85l
|
centrosomal protein 85, like |
| chr7_+_23495986 | 0.28 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr5_-_37959874 | 0.28 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr2_-_57378748 | 0.28 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr20_+_27093042 | 0.28 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr5_+_66310495 | 0.27 |
ENSDART00000114532
|
malt1
|
MALT paracaspase 1 |
| chr15_+_23657051 | 0.27 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr25_+_7532627 | 0.26 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr20_+_23408970 | 0.25 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr1_-_45614481 | 0.25 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr22_-_26274177 | 0.24 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr21_-_31252131 | 0.24 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr16_-_15988320 | 0.24 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr6_-_40899618 | 0.24 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr24_-_2829049 | 0.24 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr19_-_14155781 | 0.24 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr18_+_27571448 | 0.23 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr17_-_36529016 | 0.23 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr21_+_37090795 | 0.23 |
ENSDART00000085786
|
znf346
|
zinc finger protein 346 |
| chr7_+_28724919 | 0.22 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr3_-_49514874 | 0.21 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr13_+_8255106 | 0.21 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr17_-_36529449 | 0.19 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr21_-_2360906 | 0.19 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr7_+_62320134 | 0.18 |
ENSDART00000012089
|
stim2b
|
stromal interaction molecule 2b |
| chr7_+_28960518 | 0.17 |
ENSDART00000182903
ENSDART00000188250 ENSDART00000191830 |
dok4
|
docking protein 4 |
| chr23_-_10137322 | 0.16 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr11_-_38554394 | 0.16 |
ENSDART00000102858
|
nucks1a
|
nuclear casein kinase and cyclin-dependent kinase substrate 1a |
| chr18_-_35842554 | 0.15 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr11_-_6206520 | 0.15 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr17_-_47220934 | 0.15 |
ENSDART00000158740
|
alk
|
ALK receptor tyrosine kinase |
| chr22_+_25590391 | 0.15 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr8_+_29856013 | 0.15 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr25_+_31912153 | 0.14 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr1_+_6243906 | 0.14 |
ENSDART00000127329
|
gtf3c3
|
general transcription factor IIIC, polypeptide 3 |
| chr1_+_6646529 | 0.13 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr1_+_18811679 | 0.13 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr1_+_38858399 | 0.12 |
ENSDART00000165454
|
CU915762.1
|
|
| chr2_+_17055069 | 0.11 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr7_+_34453185 | 0.10 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr7_+_29509255 | 0.09 |
ENSDART00000076172
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr1_-_38170997 | 0.09 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr10_+_26646104 | 0.09 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr11_-_4302251 | 0.09 |
ENSDART00000182554
|
CT033790.1
|
|
| chr18_+_33788340 | 0.08 |
ENSDART00000136950
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr3_+_8826905 | 0.08 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr1_-_58505626 | 0.07 |
ENSDART00000171304
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr13_+_30421472 | 0.07 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr18_+_33762090 | 0.07 |
ENSDART00000147605
|
si:dkey-145c18.4
|
si:dkey-145c18.4 |
| chr4_+_74943111 | 0.07 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr7_+_1550966 | 0.06 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_-_37461835 | 0.06 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr23_-_44848961 | 0.05 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr25_-_35524166 | 0.04 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr8_-_2543327 | 0.03 |
ENSDART00000134115
|
si:ch211-51h9.6
|
si:ch211-51h9.6 |
| chr2_-_25120197 | 0.03 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.9 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 | 1.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 1.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.6 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.2 | 1.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 0.6 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.2 | 0.8 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.9 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 4.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.4 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.7 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 1.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 1.2 | GO:0043039 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 1.1 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.5 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.2 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 1.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 3.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 0.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 0.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 4.3 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 4.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 1.2 | GO:0032356 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 0.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 1.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 1.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 2.5 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |