Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
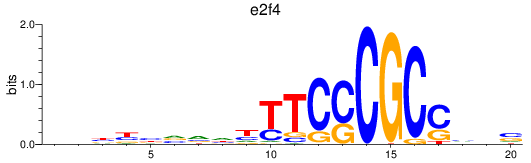
Results for e2f4
Z-value: 1.43
Transcription factors associated with e2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f4
|
ENSDARG00000101578 | E2F transcription factor 4 |
|
e2f4
|
ENSDARG00000109320 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f4 | dr11_v1_chr25_+_17405201_17405201 | 0.75 | 2.0e-02 | Click! |
Activity profile of e2f4 motif
Sorted Z-values of e2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_6884123 | 3.09 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr7_+_24814866 | 2.88 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr20_+_35445462 | 2.72 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr25_+_35155676 | 2.09 |
ENSDART00000114598
|
zgc:112234
|
zgc:112234 |
| chr3_-_7897563 | 1.99 |
ENSDART00000185232
|
ubn2b
|
ubinuclein 2b |
| chr3_-_7897305 | 1.99 |
ENSDART00000169757
|
ubn2b
|
ubinuclein 2b |
| chr5_+_20453874 | 1.90 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr25_-_36370292 | 1.84 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr7_+_5960491 | 1.82 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr23_-_19831739 | 1.73 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr20_-_29498178 | 1.66 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr11_+_31680513 | 1.54 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr23_-_45568816 | 1.54 |
ENSDART00000076086
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr15_-_23936276 | 1.52 |
ENSDART00000137569
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr7_-_6429031 | 1.46 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr7_-_50633285 | 1.42 |
ENSDART00000174018
|
crtc3
|
CREB regulated transcription coactivator 3 |
| chr7_-_73854476 | 1.37 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr10_+_6010570 | 1.36 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr7_+_24881680 | 1.27 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr20_-_28433990 | 1.24 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr7_-_6459481 | 1.24 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr17_-_11368662 | 1.21 |
ENSDART00000159061
ENSDART00000188694 ENSDART00000190932 |
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr20_+_33994580 | 1.14 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr20_-_28433616 | 1.13 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr17_-_15189397 | 1.05 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr20_-_53949798 | 1.04 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr17_+_21514777 | 1.02 |
ENSDART00000154633
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr25_-_35124576 | 1.01 |
ENSDART00000128017
|
zgc:171759
|
zgc:171759 |
| chr11_+_24348425 | 0.96 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr7_-_6415991 | 0.93 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr16_-_32128345 | 0.91 |
ENSDART00000102100
ENSDART00000137699 |
cnot3a
|
CCR4-NOT transcription complex, subunit 3a |
| chr8_+_8947623 | 0.90 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr11_-_45152702 | 0.87 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr5_+_39099380 | 0.86 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr25_-_35109536 | 0.83 |
ENSDART00000185229
|
CU302436.5
|
|
| chr1_-_20970266 | 0.82 |
ENSDART00000088545
|
klhl2
|
kelch-like family member 2 |
| chr13_+_15933168 | 0.82 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr7_-_6345507 | 0.80 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr25_+_35067318 | 0.79 |
ENSDART00000186828
|
CU302436.1
|
|
| chr14_-_6402769 | 0.79 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr13_-_45942000 | 0.79 |
ENSDART00000158534
ENSDART00000171822 ENSDART00000166442 |
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr7_+_20393386 | 0.78 |
ENSDART00000173471
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr19_-_47570672 | 0.78 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_+_30721733 | 0.76 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr19_-_48391415 | 0.72 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr10_-_38456382 | 0.71 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr25_+_35058088 | 0.68 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr25_+_35062353 | 0.65 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr18_+_39327010 | 0.63 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr2_-_57378748 | 0.61 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr12_+_47446158 | 0.58 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr2_+_37836821 | 0.56 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr10_-_37487612 | 0.54 |
ENSDART00000167964
ENSDART00000171326 |
si:dkey-65j4.2
|
si:dkey-65j4.2 |
| chr14_+_94603 | 0.53 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr16_+_35595312 | 0.53 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr13_-_22907260 | 0.51 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr13_+_48359573 | 0.49 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr22_-_26274177 | 0.46 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr24_+_41690545 | 0.44 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr23_+_20518504 | 0.41 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr3_-_16227683 | 0.40 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr24_+_17345521 | 0.38 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr3_+_22059066 | 0.31 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr3_+_29641181 | 0.26 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr7_+_34453185 | 0.24 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr22_-_21676364 | 0.21 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr20_-_33462961 | 0.14 |
ENSDART00000135927
|
si:dkey-65b13.1
|
si:dkey-65b13.1 |
| chr18_+_17020967 | 0.11 |
ENSDART00000189168
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr6_-_425378 | 0.09 |
ENSDART00000192190
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr17_-_50020672 | 0.07 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 1.4 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.2 | 2.7 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 2.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 1.5 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.2 | 1.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 3.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.2 | 0.5 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.9 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.7 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.9 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 1.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 1.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 2.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 0.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 1.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.5 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 2.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 3.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 3.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |