Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for e2f2_e2f5
Z-value: 3.30
Transcription factors associated with e2f2_e2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
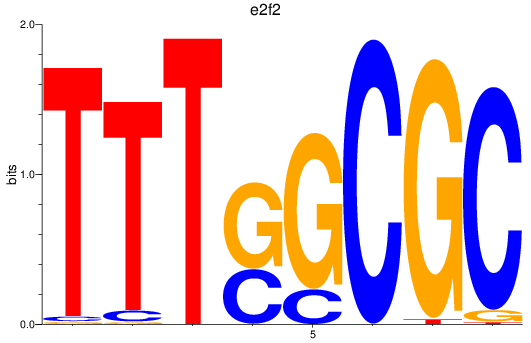
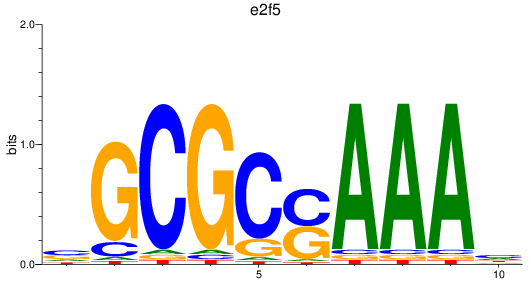
e2f2
|
ENSDARG00000079233 | si_ch211-160f23.5 |
|
e2f5
|
ENSDARG00000038812 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f5 | dr11_v1_chr2_-_31686353_31686403 | 0.88 | 2.0e-03 | Click! |
| E2F2 | dr11_v1_chr17_-_27266053_27266053 | 0.37 | 3.3e-01 | Click! |
Activity profile of e2f2_e2f5 motif
Sorted Z-values of e2f2_e2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_11792766 | 7.18 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr9_-_2892250 | 5.30 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr3_-_54607166 | 5.29 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr1_-_8553165 | 5.03 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr20_+_35438300 | 4.82 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr15_-_17099560 | 4.61 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr10_-_21362071 | 4.40 |
ENSDART00000125167
|
avd
|
avidin |
| chr2_+_44512324 | 4.15 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr7_+_55518519 | 3.84 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr9_+_22780901 | 3.79 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr13_-_13030851 | 3.69 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr22_-_4439311 | 3.66 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr2_-_29923630 | 3.52 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr2_-_29923403 | 3.51 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr22_-_38274188 | 3.44 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_26244353 | 3.39 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr6_+_33931740 | 3.32 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr7_+_41812636 | 3.19 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr9_-_2892045 | 3.18 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr7_+_41812817 | 2.94 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr25_+_35375848 | 2.84 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr7_-_6464225 | 2.80 |
ENSDART00000130760
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr19_-_2822372 | 2.79 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr5_-_54395488 | 2.78 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr5_-_12743196 | 2.74 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr19_-_20403845 | 2.72 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr2_+_38055529 | 2.68 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr19_-_47571797 | 2.67 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_+_41812190 | 2.66 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr5_+_34549845 | 2.63 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr3_-_26244256 | 2.55 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr1_-_8917902 | 2.53 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr3_+_22335030 | 2.49 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr7_-_69185124 | 2.45 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr7_-_6362460 | 2.44 |
ENSDART00000129239
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr11_+_25583950 | 2.44 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr21_+_22558187 | 2.41 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr15_+_44184367 | 2.33 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr5_-_4923473 | 2.32 |
ENSDART00000134585
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr5_+_34549365 | 2.26 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr19_-_10214264 | 2.19 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr24_+_23840821 | 2.19 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr8_+_36503797 | 2.16 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr5_+_69950882 | 2.14 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr11_+_18216404 | 2.11 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr17_+_4325693 | 2.10 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr1_+_19538299 | 2.10 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr7_+_18176162 | 2.09 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr24_-_41657005 | 2.09 |
ENSDART00000159109
|
CABZ01044099.1
|
|
| chr18_+_15271993 | 2.07 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr7_-_37895771 | 2.06 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr7_+_34236238 | 2.05 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr2_-_57941037 | 2.05 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr25_+_30196039 | 1.98 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr22_-_10591876 | 1.97 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr18_+_14329231 | 1.96 |
ENSDART00000151641
|
zgc:173742
|
zgc:173742 |
| chr6_-_34838397 | 1.95 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr8_-_1219815 | 1.92 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr12_+_17927871 | 1.92 |
ENSDART00000172062
|
trrap
|
transformation/transcription domain-associated protein |
| chr8_+_30112655 | 1.89 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr21_+_45386033 | 1.86 |
ENSDART00000151773
|
jade2
|
jade family PHD finger 2 |
| chr13_+_9612395 | 1.82 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr9_+_55455801 | 1.79 |
ENSDART00000144757
ENSDART00000186543 |
mxra5b
|
matrix-remodelling associated 5b |
| chr19_-_29294457 | 1.79 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr19_+_48060464 | 1.78 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr5_-_33255759 | 1.78 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr20_-_9199721 | 1.77 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr3_+_53116172 | 1.77 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr25_-_35047838 | 1.76 |
ENSDART00000155060
|
CU302436.3
|
|
| chr17_+_8799451 | 1.73 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr3_-_35865040 | 1.73 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr18_-_12858016 | 1.71 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr14_+_80685 | 1.71 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr13_+_8255106 | 1.68 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr17_+_26828027 | 1.67 |
ENSDART00000042060
|
jade1
|
jade family PHD finger 1 |
| chr2_-_53500424 | 1.67 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr4_-_77561679 | 1.63 |
ENSDART00000180809
|
AL935186.9
|
|
| chr13_-_14929236 | 1.60 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr7_+_71586485 | 1.60 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr21_-_233282 | 1.58 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr5_-_22969424 | 1.58 |
ENSDART00000143869
ENSDART00000172549 |
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr8_-_4760723 | 1.57 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr13_+_5013572 | 1.57 |
ENSDART00000162425
|
psap
|
prosaposin |
| chr25_-_34740627 | 1.56 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr14_-_31814149 | 1.55 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr14_-_49859747 | 1.55 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr20_+_39344889 | 1.55 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr25_+_29688671 | 1.52 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr2_+_37836821 | 1.51 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr2_-_38363017 | 1.50 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr13_-_35908275 | 1.48 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr25_+_36333993 | 1.48 |
ENSDART00000184379
|
CR354435.2
|
|
| chr16_+_28270037 | 1.47 |
ENSDART00000059035
|
mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr15_+_44201056 | 1.47 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr17_+_8799661 | 1.46 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_+_20415824 | 1.46 |
ENSDART00000009081
ENSDART00000145444 |
mob3a
|
MOB kinase activator 3A |
| chr13_+_20524921 | 1.46 |
ENSDART00000081385
|
ccdc172
|
coiled-coil domain containing 172 |
| chr1_-_59216197 | 1.45 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr20_-_29499363 | 1.45 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_-_73845736 | 1.43 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr3_-_15679107 | 1.43 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr3_+_37112693 | 1.43 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr2_+_8779164 | 1.43 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr19_-_20403318 | 1.41 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr5_+_58679071 | 1.38 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr21_-_19915757 | 1.37 |
ENSDART00000164317
|
eri1
|
exoribonuclease 1 |
| chr17_-_26537928 | 1.34 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr22_-_5822147 | 1.33 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr18_+_41560822 | 1.30 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr19_-_2582858 | 1.28 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr7_+_24814866 | 1.28 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr20_-_44576949 | 1.27 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr25_-_35144620 | 1.26 |
ENSDART00000185669
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr25_-_13403726 | 1.25 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr18_+_41561285 | 1.25 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr13_+_33462232 | 1.25 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr10_+_21434649 | 1.25 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr13_+_31402067 | 1.25 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr16_+_11188810 | 1.23 |
ENSDART00000186011
|
cicb
|
capicua transcriptional repressor b |
| chr8_-_24791060 | 1.23 |
ENSDART00000111617
|
rbm15
|
RNA binding motif protein 15 |
| chr20_+_41801913 | 1.23 |
ENSDART00000139805
|
mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr12_+_9183626 | 1.22 |
ENSDART00000020192
|
tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr13_-_35907768 | 1.21 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr14_-_35400671 | 1.20 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr24_-_37338739 | 1.20 |
ENSDART00000146844
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr4_-_15003854 | 1.18 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr2_-_44777592 | 1.18 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr10_-_20588787 | 1.18 |
ENSDART00000138045
ENSDART00000181885 ENSDART00000091115 |
nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr25_-_35095129 | 1.15 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr21_-_19916021 | 1.14 |
ENSDART00000065671
|
eri1
|
exoribonuclease 1 |
| chr17_-_33707589 | 1.14 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr20_-_38827623 | 1.14 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_71535045 | 1.14 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr3_-_34528306 | 1.13 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr20_-_29498178 | 1.13 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr14_-_30918662 | 1.13 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr5_+_20453874 | 1.12 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr16_-_43335914 | 1.12 |
ENSDART00000111963
|
atad2
|
ATPase family, AAA domain containing 2 |
| chr2_-_44344321 | 1.11 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr25_-_35140746 | 1.10 |
ENSDART00000129969
|
si:ch211-113a14.19
|
si:ch211-113a14.19 |
| chr23_-_45339439 | 1.09 |
ENSDART00000148726
|
ccdc171
|
coiled-coil domain containing 171 |
| chr4_+_18441988 | 1.09 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr7_+_19851422 | 1.09 |
ENSDART00000142970
ENSDART00000190027 |
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr23_-_17003533 | 1.09 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr14_-_31715373 | 1.08 |
ENSDART00000127303
ENSDART00000173274 ENSDART00000173435 ENSDART00000172876 ENSDART00000173036 |
map7d3
|
MAP7 domain containing 3 |
| chr24_-_11908115 | 1.08 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr25_-_32363341 | 1.07 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr16_+_41015781 | 1.07 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr5_+_12743640 | 1.06 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr17_-_22573311 | 1.06 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr20_-_33487729 | 1.06 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr17_+_18810904 | 1.05 |
ENSDART00000130899
|
si:dkey-288a3.2
|
si:dkey-288a3.2 |
| chr21_-_30032706 | 1.05 |
ENSDART00000156721
|
pwwp2a
|
PWWP domain containing 2A |
| chr2_-_48375342 | 1.04 |
ENSDART00000148788
|
per2
|
period circadian clock 2 |
| chr2_+_11119315 | 1.04 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr12_-_33817114 | 1.04 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr4_+_69888328 | 1.04 |
ENSDART00000170163
|
si:ch211-145h19.5
|
si:ch211-145h19.5 |
| chr6_+_50451337 | 1.04 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr20_+_38201644 | 1.03 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr18_-_20532339 | 1.03 |
ENSDART00000060291
|
ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr5_-_20491198 | 0.99 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr7_+_5956937 | 0.99 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr19_-_20777351 | 0.99 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr25_-_35145920 | 0.99 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr19_-_47571456 | 0.99 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_+_27690 | 0.98 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr7_+_5908077 | 0.96 |
ENSDART00000173404
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr21_-_30031396 | 0.95 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr21_+_233271 | 0.95 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr7_+_24023653 | 0.95 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr15_-_1584137 | 0.94 |
ENSDART00000056772
|
trim59
|
tripartite motif containing 59 |
| chr2_+_21000334 | 0.93 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr7_+_67748939 | 0.93 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr7_-_60351876 | 0.93 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr21_-_13055195 | 0.93 |
ENSDART00000133517
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr18_-_46183462 | 0.92 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr10_+_9595575 | 0.92 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr12_-_6880694 | 0.90 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr15_-_7337148 | 0.90 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr7_-_40657831 | 0.90 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr5_+_51248784 | 0.90 |
ENSDART00000159571
ENSDART00000189513 ENSDART00000092285 |
msh3
|
mutS homolog 3 (E. coli) |
| chr9_+_29431763 | 0.90 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr8_-_25761544 | 0.89 |
ENSDART00000078152
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr8_+_36509885 | 0.89 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr24_+_17345521 | 0.88 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr1_+_26677406 | 0.88 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr20_+_44582318 | 0.88 |
ENSDART00000149000
ENSDART00000149775 ENSDART00000085416 |
atad2b
|
ATPase family, AAA domain containing 2B |
| chr7_-_7797654 | 0.87 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr19_-_4840688 | 0.87 |
ENSDART00000193855
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr7_-_73854476 | 0.86 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr15_-_4124099 | 0.86 |
ENSDART00000185583
|
rnf168
|
ring finger protein 168 |
| chr21_-_40562705 | 0.85 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr25_-_35112466 | 0.85 |
ENSDART00000183481
|
CU302436.4
|
|
| chr5_+_27488975 | 0.85 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr6_-_33931696 | 0.84 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| chr13_+_46803979 | 0.84 |
ENSDART00000159260
|
CU695232.1
|
|
| chr23_+_45339684 | 0.84 |
ENSDART00000149410
ENSDART00000102441 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr4_-_5831522 | 0.83 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr9_+_22466218 | 0.82 |
ENSDART00000131214
|
dgkg
|
diacylglycerol kinase, gamma |
| chr1_+_9199031 | 0.82 |
ENSDART00000092058
ENSDART00000182771 |
chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f2_e2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 1.3 | 10.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.3 | 6.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 1.2 | 3.7 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.9 | 0.9 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.8 | 5.9 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.8 | 3.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.7 | 2.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.7 | 4.1 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.7 | 7.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 1.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.6 | 5.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.6 | 1.8 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.6 | 2.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 1.7 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.5 | 1.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.5 | 4.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 2.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 1.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.4 | 2.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.4 | 4.8 | GO:0030719 | P granule organization(GO:0030719) |
| 0.4 | 3.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.4 | 3.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.4 | 1.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.3 | 1.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 2.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 0.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.3 | 2.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 1.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 4.9 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 0.7 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 8.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.2 | 1.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 2.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 1.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 1.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 3.3 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.2 | 0.8 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 3.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 1.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.9 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 1.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 10.5 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.2 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.8 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 3.0 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.6 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 1.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 2.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.9 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 2.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 2.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.6 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.2 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 2.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 2.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.7 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.8 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 1.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.9 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 3.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 1.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 3.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.7 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.8 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.5 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 1.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0030643 | microglia differentiation(GO:0014004) cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.9 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 9.7 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 2.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.7 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.7 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 3.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.8 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 1.8 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.9 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 2.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.8 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 1.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.6 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.7 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.7 | GO:0031929 | TOR signaling(GO:0031929) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 12.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.6 | 4.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 2.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 2.5 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.4 | 2.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 1.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.3 | 1.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 3.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 1.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.8 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 2.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 4.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.5 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.8 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 2.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 2.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 7.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 4.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 10.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.2 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 1.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.2 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.9 | 6.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 4.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 1.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.5 | 6.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 4.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 1.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 10.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.6 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.4 | 1.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 1.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 2.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.4 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 1.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 2.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 4.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 0.8 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 0.8 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.3 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 2.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 0.9 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 2.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 2.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 1.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.8 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 0.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 1.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 2.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.4 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 2.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.5 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 1.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 2.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 13.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.9 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 2.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 2.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 4.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.8 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.3 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 9.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 10.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 6.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 5.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 15.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 2.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.9 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.9 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.1 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.8 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |