Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
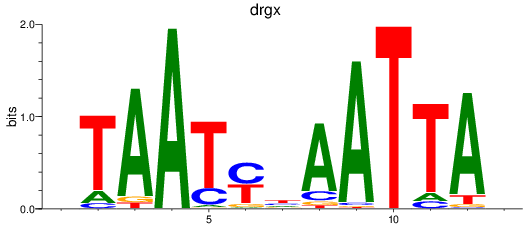
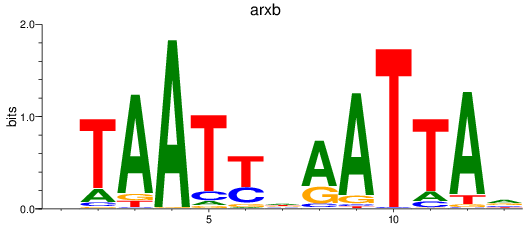
Results for drgx_arxb
Z-value: 1.26
Transcription factors associated with drgx_arxb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
drgx
|
ENSDARG00000069329 | dorsal root ganglia homeobox |
|
arxb
|
ENSDARG00000101523 | aristaless related homeobox b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| drgx | dr11_v1_chr13_+_30912385_30912385 | 0.88 | 2.0e-03 | Click! |
| arxb | dr11_v1_chr21_-_40676224_40676224 | 0.61 | 8.0e-02 | Click! |
Activity profile of drgx_arxb motif
Sorted Z-values of drgx_arxb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 11.01 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr21_+_7582036 | 3.23 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr4_+_9669717 | 3.00 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr7_+_26629084 | 2.26 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr9_+_34641237 | 2.23 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr10_-_26744131 | 2.06 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr16_-_31717851 | 2.04 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr6_+_23887314 | 1.94 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr12_+_42436920 | 1.85 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr16_-_31718013 | 1.73 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr25_-_13381854 | 1.71 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr19_+_40856534 | 1.58 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr22_-_32507966 | 1.52 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr24_-_38384432 | 1.50 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr2_+_30916188 | 1.42 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr19_+_9174166 | 1.39 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr1_-_40911332 | 1.37 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr20_-_43775495 | 1.35 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr13_-_31622195 | 1.34 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr25_+_18964782 | 1.28 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr9_-_1959917 | 1.23 |
ENSDART00000082359
|
hoxd3a
|
homeobox D3a |
| chr14_-_17068511 | 1.22 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr17_+_18117029 | 1.20 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr19_+_40856807 | 1.15 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr21_-_44104600 | 1.08 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr13_-_29421331 | 1.06 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr2_+_37227011 | 1.06 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr21_-_17482465 | 1.00 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr20_-_30920356 | 0.99 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr23_-_24682244 | 0.98 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr7_-_28148310 | 0.98 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr7_+_66822229 | 0.97 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr21_-_27881752 | 0.96 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr4_-_14315855 | 0.94 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr3_-_32337653 | 0.93 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr16_+_23984755 | 0.92 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr12_+_42436328 | 0.89 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr23_-_12345764 | 0.84 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr18_+_5490668 | 0.82 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr12_-_20795867 | 0.81 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr14_-_17068712 | 0.79 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr3_+_29445473 | 0.74 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr20_+_34320635 | 0.74 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr13_+_1726953 | 0.74 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr6_+_40661703 | 0.74 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr10_-_34772211 | 0.74 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr17_+_30545895 | 0.71 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr24_-_7697274 | 0.70 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr6_+_37894220 | 0.70 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr9_-_46701326 | 0.67 |
ENSDART00000159971
|
si:ch73-193i22.1
|
si:ch73-193i22.1 |
| chr9_-_51436377 | 0.67 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr20_+_31287356 | 0.65 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr6_+_9130989 | 0.63 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr16_+_22654481 | 0.61 |
ENSDART00000179762
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr10_+_32104305 | 0.61 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr14_+_19258702 | 0.59 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr8_-_7391721 | 0.59 |
ENSDART00000149836
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr24_+_25069609 | 0.59 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr19_+_10339538 | 0.58 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr15_+_19797918 | 0.58 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr15_-_17813680 | 0.58 |
ENSDART00000158556
|
CT573342.2
|
|
| chr15_-_17868870 | 0.55 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr18_+_38321039 | 0.54 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr1_+_25801648 | 0.54 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr21_-_20733615 | 0.52 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr15_-_17869115 | 0.51 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr10_-_33621739 | 0.50 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr25_-_19648154 | 0.45 |
ENSDART00000148570
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr1_+_53954230 | 0.45 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr15_+_45640906 | 0.45 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr18_-_26785861 | 0.43 |
ENSDART00000098361
|
nmba
|
neuromedin Ba |
| chr4_-_16001118 | 0.42 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr5_+_34407763 | 0.42 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr20_-_20931197 | 0.42 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr18_+_6054816 | 0.41 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr23_+_28770225 | 0.40 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr5_+_11407504 | 0.39 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr12_+_17504559 | 0.39 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr17_-_26926577 | 0.39 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr15_-_21702317 | 0.39 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr20_-_20930926 | 0.38 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr5_-_58840971 | 0.38 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr20_-_2134620 | 0.36 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr22_+_17784414 | 0.34 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr10_-_1725699 | 0.33 |
ENSDART00000183555
|
gal3st1b
|
galactose-3-O-sulfotransferase 1b |
| chr7_+_20344222 | 0.33 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr4_+_2267641 | 0.32 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr7_+_20147202 | 0.32 |
ENSDART00000011398
|
si:ch73-335l21.1
|
si:ch73-335l21.1 |
| chr5_+_48683447 | 0.31 |
ENSDART00000008043
|
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr24_+_21346796 | 0.31 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr2_-_30135446 | 0.30 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr17_-_19345521 | 0.29 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr21_+_19008168 | 0.28 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr6_+_49081141 | 0.27 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr3_-_12970418 | 0.27 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr14_-_21123551 | 0.26 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr6_+_6924637 | 0.24 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr21_+_10739846 | 0.24 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr2_-_5505094 | 0.24 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr21_-_43022048 | 0.23 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr7_-_30367650 | 0.23 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr16_-_27640995 | 0.22 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr7_-_52334840 | 0.21 |
ENSDART00000174173
|
CR938716.1
|
|
| chr2_+_20604775 | 0.19 |
ENSDART00000131501
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr10_-_5581487 | 0.18 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr14_-_29826659 | 0.17 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr10_-_32920690 | 0.17 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr15_+_23550890 | 0.17 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr11_-_29816429 | 0.16 |
ENSDART00000069132
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr10_+_29698467 | 0.15 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr21_-_10446405 | 0.14 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr6_-_3573716 | 0.14 |
ENSDART00000026693
|
dmbx1b
|
diencephalon/mesencephalon homeobox 1b |
| chr3_-_16493528 | 0.14 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr2_+_49417900 | 0.14 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr6_+_58698475 | 0.11 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr5_-_30481263 | 0.11 |
ENSDART00000086734
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr11_-_37425407 | 0.11 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr12_-_32013125 | 0.11 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr14_-_22152064 | 0.10 |
ENSDART00000115422
|
nudcd2
|
NudC domain containing 2 |
| chr11_-_45138857 | 0.10 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr9_-_27649406 | 0.10 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr12_+_10631266 | 0.10 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr18_+_33725576 | 0.09 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr2_-_20890429 | 0.08 |
ENSDART00000149989
|
pdca
|
phosducin a |
| chr15_+_31332552 | 0.08 |
ENSDART00000134933
ENSDART00000173915 |
or119-2
|
odorant receptor, family F, subfamily 119, member 2 |
| chr3_+_46628885 | 0.08 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr14_-_46070802 | 0.08 |
ENSDART00000038670
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr24_-_38079261 | 0.08 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr4_-_28158335 | 0.07 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr5_+_40224938 | 0.07 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr14_-_29859067 | 0.07 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr3_-_13921173 | 0.06 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr9_-_21825913 | 0.06 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr24_-_38657683 | 0.06 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr23_-_1658980 | 0.06 |
ENSDART00000186227
|
CU693481.1
|
|
| chr1_-_46706639 | 0.06 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr22_+_21324398 | 0.06 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr17_-_12966907 | 0.05 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr5_+_42400777 | 0.05 |
ENSDART00000183114
|
BX548073.8
|
|
| chr7_+_20344032 | 0.05 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr6_+_8315050 | 0.05 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr17_-_39772999 | 0.04 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr22_-_15693085 | 0.04 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr7_+_21859337 | 0.03 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr12_-_19091214 | 0.03 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr1_+_18811679 | 0.03 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr1_-_49250490 | 0.03 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr1_-_55873178 | 0.02 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr8_-_21372446 | 0.02 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr2_-_42071558 | 0.02 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr7_+_41303572 | 0.02 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr5_-_68074592 | 0.02 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr21_-_18648861 | 0.01 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr2_+_20605186 | 0.01 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr23_-_16484383 | 0.01 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr15_+_8767650 | 0.00 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of drgx_arxb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0021767 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.5 | 2.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 1.3 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.3 | 1.5 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 11.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.6 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 2.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 1.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.7 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 1.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.3 | GO:0050960 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.1 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 2.2 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.4 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 1.4 | GO:0006936 | muscle contraction(GO:0006936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 2.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 1.3 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 2.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.7 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |