Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
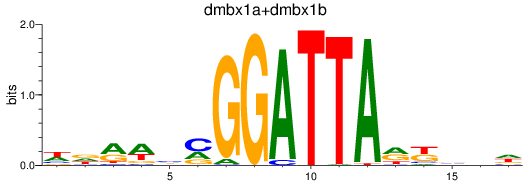
Results for dmbx1a+dmbx1b
Z-value: 0.61
Transcription factors associated with dmbx1a+dmbx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dmbx1b
|
ENSDARG00000002510 | diencephalon/mesencephalon homeobox 1b |
|
dmbx1a
|
ENSDARG00000009922 | diencephalon/mesencephalon homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dmbx1b | dr11_v1_chr6_-_3573716_3573716 | -0.36 | 3.5e-01 | Click! |
| dmbx1a | dr11_v1_chr2_-_10192459_10192487 | -0.17 | 6.6e-01 | Click! |
Activity profile of dmbx1a+dmbx1b motif
Sorted Z-values of dmbx1a+dmbx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_56577906 | 0.93 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr4_-_16628801 | 0.46 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr18_+_20567542 | 0.45 |
ENSDART00000182585
|
bida
|
BH3 interacting domain death agonist |
| chr1_-_12126535 | 0.40 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr23_-_3759692 | 0.38 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr12_+_33894396 | 0.38 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr23_-_3759345 | 0.37 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr13_-_31938512 | 0.35 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr3_+_23092762 | 0.32 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr20_-_42534504 | 0.28 |
ENSDART00000132310
|
rfx6
|
regulatory factor X, 6 |
| chr23_-_545942 | 0.27 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr19_-_30811161 | 0.27 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr18_-_10713414 | 0.26 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr15_-_6976851 | 0.25 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr5_-_22602979 | 0.25 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr3_-_41995321 | 0.25 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr12_+_33894665 | 0.25 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr14_-_46238186 | 0.25 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr18_-_10713230 | 0.24 |
ENSDART00000183646
|
rbm28
|
RNA binding motif protein 28 |
| chr19_+_2631565 | 0.23 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr16_-_46578523 | 0.23 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr5_-_22602780 | 0.23 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr12_+_17042754 | 0.22 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr2_-_9744081 | 0.22 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr25_-_35121946 | 0.22 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr19_+_24324967 | 0.21 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr4_+_8680767 | 0.21 |
ENSDART00000182726
|
adipor2
|
adiponectin receptor 2 |
| chr16_+_16265850 | 0.21 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr15_-_7337148 | 0.21 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr4_-_59152337 | 0.20 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr1_-_9130136 | 0.20 |
ENSDART00000113013
|
palb2
|
partner and localizer of BRCA2 |
| chr5_-_69180227 | 0.20 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr2_+_44972720 | 0.20 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr11_+_29671661 | 0.20 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr5_-_25733745 | 0.19 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr4_-_76488581 | 0.19 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr7_-_21905851 | 0.19 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr8_+_46327350 | 0.18 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr10_-_7988396 | 0.18 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr9_+_48123224 | 0.17 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr19_+_3140313 | 0.17 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr10_+_6013076 | 0.16 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr17_+_10593398 | 0.16 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr23_-_1065435 | 0.15 |
ENSDART00000138150
|
si:ch1073-272j17.3
|
si:ch1073-272j17.3 |
| chr4_+_55593843 | 0.15 |
ENSDART00000163356
|
znf1078
|
zinc finger protein 1078 |
| chr5_-_5243079 | 0.14 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr13_+_1015749 | 0.14 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr2_+_243778 | 0.13 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr23_-_27050083 | 0.13 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr5_-_69180587 | 0.12 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr4_-_4256300 | 0.12 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr4_-_42039794 | 0.12 |
ENSDART00000110313
|
si:dkeyp-85d8.5
|
si:dkeyp-85d8.5 |
| chr18_+_50525109 | 0.12 |
ENSDART00000098390
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr4_+_47385355 | 0.12 |
ENSDART00000163413
|
znf1122
|
zinc finger protein 1122 |
| chr7_+_65121743 | 0.12 |
ENSDART00000108471
|
ipo7
|
importin 7 |
| chr23_-_29553430 | 0.12 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr13_-_14269626 | 0.12 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr2_-_38363017 | 0.12 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr12_+_33460794 | 0.12 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr4_-_50722304 | 0.11 |
ENSDART00000150480
|
znf1110
|
zinc finger protein 1110 |
| chr16_-_21692024 | 0.11 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr17_-_21617618 | 0.11 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr23_+_16815353 | 0.11 |
ENSDART00000142789
ENSDART00000143424 |
zgc:100832
|
zgc:100832 |
| chr25_+_36411624 | 0.10 |
ENSDART00000124347
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr20_-_21672970 | 0.10 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr8_+_54263946 | 0.10 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr16_+_37876779 | 0.10 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr17_-_8656155 | 0.09 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr10_+_21899753 | 0.09 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr23_-_29553691 | 0.09 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr4_+_33012407 | 0.09 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr4_+_31450683 | 0.08 |
ENSDART00000169303
|
znf996
|
zinc finger protein 996 |
| chr15_-_41734639 | 0.08 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr4_-_56083825 | 0.08 |
ENSDART00000157710
|
znf986
|
zinc finger protein 986 |
| chr6_-_39653972 | 0.08 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr4_+_36906436 | 0.08 |
ENSDART00000136773
|
BX537137.1
|
|
| chr13_-_35892243 | 0.08 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr13_-_36581875 | 0.08 |
ENSDART00000113204
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr5_-_8817841 | 0.07 |
ENSDART00000170921
|
fgf10b
|
fibroblast growth factor 10b |
| chr1_+_35949269 | 0.07 |
ENSDART00000180628
|
BX276105.3
|
|
| chr18_+_21951551 | 0.07 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr20_-_33515890 | 0.07 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr23_+_16814968 | 0.07 |
ENSDART00000161299
|
zgc:100832
|
zgc:100832 |
| chr9_+_21990095 | 0.06 |
ENSDART00000146829
ENSDART00000133515 ENSDART00000193582 |
si:dkey-57a22.13
|
si:dkey-57a22.13 |
| chr7_-_54320088 | 0.06 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr18_+_33172634 | 0.06 |
ENSDART00000151519
|
si:ch73-125k17.2
|
si:ch73-125k17.2 |
| chr8_+_14381272 | 0.06 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr4_-_18455480 | 0.06 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr11_+_27973055 | 0.05 |
ENSDART00000135135
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr20_+_12830448 | 0.05 |
ENSDART00000164754
|
BX547930.4
|
|
| chr10_-_18468385 | 0.05 |
ENSDART00000183302
|
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr13_-_35892051 | 0.04 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_19904136 | 0.04 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr25_-_32381642 | 0.04 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr18_-_21950751 | 0.04 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr22_+_2709467 | 0.04 |
ENSDART00000141416
|
znf1171
|
zinc finger protein 1171 |
| chr4_+_73876128 | 0.04 |
ENSDART00000174225
|
CABZ01069022.1
|
|
| chr7_-_35409027 | 0.04 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr3_-_21402279 | 0.03 |
ENSDART00000164513
|
CT573446.1
|
|
| chr23_-_26652273 | 0.03 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr4_-_49133107 | 0.03 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr14_-_3944017 | 0.03 |
ENSDART00000055817
|
si:ch73-49o8.1
|
si:ch73-49o8.1 |
| chr25_+_11456696 | 0.03 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr21_+_44773237 | 0.02 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr14_+_2487672 | 0.02 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr5_+_25733774 | 0.02 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr5_-_8817458 | 0.02 |
ENSDART00000191098
|
fgf10b
|
fibroblast growth factor 10b |
| chr12_+_292452 | 0.02 |
ENSDART00000189944
|
CABZ01058222.1
|
|
| chr22_+_13886821 | 0.02 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr7_+_19903924 | 0.02 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr20_-_31308805 | 0.02 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
| chr23_+_45512825 | 0.02 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr3_-_15139293 | 0.02 |
ENSDART00000161213
ENSDART00000146053 |
fam173a
|
family with sequence similarity 173, member A |
| chr12_-_13081229 | 0.02 |
ENSDART00000152510
ENSDART00000126866 |
ccser2b
|
coiled-coil serine-rich protein 2b |
| chr18_+_5172848 | 0.02 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr4_+_40383229 | 0.01 |
ENSDART00000126906
ENSDART00000191198 |
si:ch211-218h8.1
|
si:ch211-218h8.1 |
| chr4_-_76488854 | 0.01 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr25_+_19041329 | 0.01 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr21_+_21709204 | 0.01 |
ENSDART00000137478
|
or125-1
|
odorant receptor, family E, subfamily 125, member 1 |
| chr3_+_34919810 | 0.01 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr1_+_52560549 | 0.01 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr22_+_6740039 | 0.00 |
ENSDART00000144122
|
CT583625.2
|
|
| chr18_+_15182965 | 0.00 |
ENSDART00000137018
|
si:ch73-62b13.1
|
si:ch73-62b13.1 |
| chr18_-_5103931 | 0.00 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
Network of associatons between targets according to the STRING database.
First level regulatory network of dmbx1a+dmbx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.3 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0044803 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.2 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.1 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.3 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |