Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for dlx5a
Z-value: 0.40
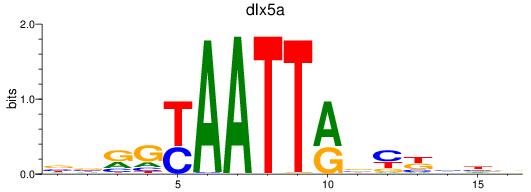
Transcription factors associated with dlx5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx5a
|
ENSDARG00000042296 | distal-less homeobox 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx5a | dr11_v1_chr19_-_41472228_41472228 | -0.82 | 7.4e-03 | Click! |
Activity profile of dlx5a motif
Sorted Z-values of dlx5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_19596214 | 0.89 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr2_+_6253246 | 0.75 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr4_+_306036 | 0.55 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr24_+_1023839 | 0.52 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr11_-_44801968 | 0.52 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr10_-_35149513 | 0.43 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr6_-_43283122 | 0.40 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr13_-_33227411 | 0.30 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr17_+_43867889 | 0.30 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr23_-_31913069 | 0.30 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_43863708 | 0.30 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr16_-_42056137 | 0.29 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_-_14114078 | 0.29 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr2_-_21819421 | 0.28 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_11290828 | 0.28 |
ENSDART00000184078
|
rlim
|
ring finger protein, LIM domain interacting |
| chr20_+_46513651 | 0.24 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr11_+_31864921 | 0.24 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr6_+_23026714 | 0.23 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr8_-_39822917 | 0.22 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr2_-_2451181 | 0.22 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr16_+_35661771 | 0.22 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr3_-_23643751 | 0.21 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr17_+_49500820 | 0.21 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr23_-_31913231 | 0.21 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_-_51368681 | 0.20 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr22_-_11517377 | 0.19 |
ENSDART00000193885
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr17_-_43863700 | 0.19 |
ENSDART00000157530
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr15_+_29140126 | 0.19 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr6_-_40922971 | 0.18 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr16_+_35661331 | 0.17 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr16_+_42471455 | 0.17 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr20_+_14114258 | 0.17 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr19_-_27336092 | 0.16 |
ENSDART00000176885
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr7_-_12464412 | 0.15 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr3_+_32832538 | 0.14 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr23_+_34990693 | 0.14 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr1_+_40199074 | 0.13 |
ENSDART00000179679
ENSDART00000136849 |
si:ch211-113e8.5
|
si:ch211-113e8.5 |
| chr16_+_28994709 | 0.13 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr1_-_53277413 | 0.13 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr10_+_20392083 | 0.13 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr10_-_13343831 | 0.12 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr3_-_23512285 | 0.12 |
ENSDART00000159151
|
BX682558.1
|
|
| chr12_+_1469090 | 0.11 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr12_-_14211293 | 0.11 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr19_+_20793388 | 0.11 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr16_+_45425181 | 0.10 |
ENSDART00000168591
|
phf1
|
PHD finger protein 1 |
| chr1_-_13989643 | 0.10 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr19_+_20793237 | 0.10 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr25_-_7705487 | 0.10 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr8_-_537716 | 0.09 |
ENSDART00000051777
|
zgc:101664
|
zgc:101664 |
| chr20_-_8094718 | 0.09 |
ENSDART00000122902
|
si:ch211-232i5.3
|
si:ch211-232i5.3 |
| chr7_+_69019851 | 0.09 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr9_-_49964810 | 0.09 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_-_63848305 | 0.09 |
ENSDART00000097308
|
BX901974.1
|
|
| chr10_+_2529037 | 0.09 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr17_+_25187226 | 0.08 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr24_+_7336411 | 0.08 |
ENSDART00000191170
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr7_-_69647988 | 0.08 |
ENSDART00000169943
|
LO018231.1
|
|
| chr24_-_39637418 | 0.07 |
ENSDART00000142522
|
fam234a
|
family with sequence similarity 234, member A |
| chr18_+_50461981 | 0.07 |
ENSDART00000158761
|
CU896640.1
|
|
| chr14_-_14705750 | 0.07 |
ENSDART00000168092
|
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr23_+_44374041 | 0.06 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr20_+_35484070 | 0.05 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr16_-_35329803 | 0.05 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr24_-_32582378 | 0.05 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr8_-_50888806 | 0.04 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr16_+_45424907 | 0.04 |
ENSDART00000134471
|
phf1
|
PHD finger protein 1 |
| chr23_+_31912882 | 0.04 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr4_-_5795309 | 0.04 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr4_-_76184243 | 0.04 |
ENSDART00000180937
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr20_+_39457598 | 0.04 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr2_+_38370535 | 0.03 |
ENSDART00000179614
|
psmb11a
|
proteasome subunit beta 11a |
| chr9_+_13229585 | 0.03 |
ENSDART00000154879
ENSDART00000141705 |
carf
|
calcium responsive transcription factor |
| chr13_+_24829996 | 0.03 |
ENSDART00000147194
|
si:ch211-223p8.8
|
si:ch211-223p8.8 |
| chr20_-_3004126 | 0.03 |
ENSDART00000064390
|
ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr21_+_39948300 | 0.02 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr1_-_17715493 | 0.02 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr23_+_33963619 | 0.02 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr16_-_30934319 | 0.02 |
ENSDART00000109940
|
gpr20
|
G protein-coupled receptor 20 |
| chr19_+_5480327 | 0.01 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr16_-_31525290 | 0.00 |
ENSDART00000109996
|
wisp1b
|
WNT1 inducible signaling pathway protein 1b |
| chr5_+_66433287 | 0.00 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0048798 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |