Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
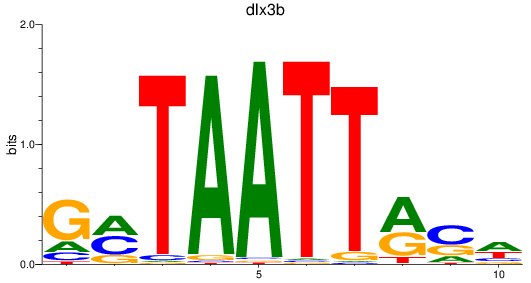
Results for dlx3b
Z-value: 1.03
Transcription factors associated with dlx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx3b
|
ENSDARG00000014626 | distal-less homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx3b | dr11_v1_chr12_+_5708400_5708400 | 0.69 | 4.1e-02 | Click! |
Activity profile of dlx3b motif
Sorted Z-values of dlx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_42607451 | 2.13 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr14_-_36378494 | 2.02 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr23_+_36130883 | 1.98 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr15_+_47903864 | 1.78 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr10_+_32104305 | 1.72 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr11_-_6974022 | 1.66 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr14_-_24410673 | 1.44 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr16_+_5774977 | 1.41 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr25_+_29160102 | 1.35 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr11_-_25418856 | 1.33 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr18_+_9171778 | 1.26 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr21_+_6751405 | 1.25 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr14_+_36220479 | 1.25 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr1_-_58036509 | 1.23 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr21_+_6751760 | 1.23 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr5_+_56268436 | 1.21 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr3_-_46811611 | 1.17 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_-_34044369 | 1.16 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_1185772 | 1.15 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr14_-_30587814 | 1.15 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr4_-_9891874 | 1.14 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr16_+_31804590 | 1.12 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr3_+_23743139 | 1.12 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr24_+_22485710 | 1.09 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr3_+_23742868 | 1.08 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr13_+_35339182 | 1.08 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr11_+_30244356 | 1.05 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr16_-_20312146 | 1.04 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr7_-_25895189 | 1.04 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr11_-_39105253 | 1.03 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr20_-_9462433 | 0.98 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr23_+_36144487 | 0.97 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr7_-_54679595 | 0.96 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr4_-_17629444 | 0.94 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr11_+_38280454 | 0.92 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr5_-_51619742 | 0.91 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr17_-_15546862 | 0.90 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr4_-_15420452 | 0.90 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr4_+_12612145 | 0.89 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr24_-_17029374 | 0.88 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr8_-_15129573 | 0.84 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr3_-_28428198 | 0.83 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_+_30787903 | 0.82 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr4_+_41311345 | 0.81 |
ENSDART00000151905
|
si:dkey-16p19.8
|
si:dkey-16p19.8 |
| chr6_+_39370587 | 0.81 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr20_+_6659770 | 0.80 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr19_+_22062202 | 0.79 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr5_-_48268049 | 0.78 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr18_-_46258612 | 0.77 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr23_-_7799184 | 0.74 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr2_-_23004286 | 0.74 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr2_+_35612621 | 0.72 |
ENSDART00000143082
|
si:dkey-4i23.5
|
si:dkey-4i23.5 |
| chr7_+_15872357 | 0.71 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr15_-_42736433 | 0.70 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr17_-_20717845 | 0.70 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr24_-_17023392 | 0.67 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr1_+_23783349 | 0.67 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr20_-_27225876 | 0.67 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr20_-_32270866 | 0.65 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr19_-_32641725 | 0.64 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr10_+_22381802 | 0.63 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr23_+_28582865 | 0.60 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr14_-_33821515 | 0.59 |
ENSDART00000173218
|
vimr2
|
vimentin-related 2 |
| chr3_-_21280373 | 0.59 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr23_-_20051369 | 0.59 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr17_+_44780166 | 0.57 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr11_-_42554290 | 0.57 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr10_-_1788376 | 0.56 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr9_+_11532025 | 0.56 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr20_+_4060839 | 0.56 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr1_-_19402802 | 0.55 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr6_-_50203682 | 0.55 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr8_-_23776399 | 0.54 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr4_+_14727018 | 0.54 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr6_-_6487876 | 0.54 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr4_+_3980247 | 0.51 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_+_18588551 | 0.51 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr23_+_26946744 | 0.51 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr25_-_18913336 | 0.50 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr22_+_20427170 | 0.50 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr23_+_43489182 | 0.48 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr21_-_37790727 | 0.48 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr8_-_31107537 | 0.48 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr1_-_44701313 | 0.48 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr2_+_21229909 | 0.47 |
ENSDART00000046127
|
vipr1a
|
vasoactive intestinal peptide receptor 1a |
| chr17_+_21295132 | 0.47 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr12_+_22580579 | 0.46 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr7_+_30926289 | 0.45 |
ENSDART00000173518
|
si:dkey-1h4.4
|
si:dkey-1h4.4 |
| chr25_-_3470910 | 0.44 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr9_+_21165484 | 0.44 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr19_+_7735157 | 0.44 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr21_-_26490186 | 0.44 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr20_-_35578435 | 0.43 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr9_-_19161982 | 0.42 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr20_+_37294112 | 0.42 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr3_+_17537352 | 0.41 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr4_+_12612723 | 0.41 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr6_-_35472923 | 0.41 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr4_+_14727212 | 0.41 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr22_-_24284447 | 0.41 |
ENSDART00000149894
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chrM_+_9052 | 0.39 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr4_+_57580303 | 0.38 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr21_-_5856050 | 0.37 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr19_-_19871211 | 0.37 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr20_+_30490682 | 0.35 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr8_-_24252933 | 0.34 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr8_+_48484455 | 0.33 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr9_-_32730487 | 0.33 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr4_+_49322310 | 0.32 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr14_-_17622080 | 0.30 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr24_+_35881124 | 0.30 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr21_+_15824182 | 0.27 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr21_+_11244068 | 0.26 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr15_+_5360407 | 0.24 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr18_-_2433011 | 0.23 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr16_-_21620947 | 0.23 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_17235891 | 0.23 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr22_+_21324398 | 0.23 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr16_-_13623928 | 0.22 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr10_+_3049636 | 0.22 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr6_+_24398907 | 0.21 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr23_-_19230627 | 0.21 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr21_+_30355767 | 0.21 |
ENSDART00000189948
|
CR749164.1
|
|
| chr10_+_21867307 | 0.19 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr23_-_18130264 | 0.19 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr20_+_23440632 | 0.17 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr16_-_19890303 | 0.17 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr17_-_29192987 | 0.16 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr15_-_9272328 | 0.16 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr22_-_14262115 | 0.15 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr19_+_46158078 | 0.15 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr9_-_23152092 | 0.15 |
ENSDART00000180155
ENSDART00000186935 |
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr24_-_29963858 | 0.14 |
ENSDART00000183442
|
CR352310.1
|
|
| chr25_+_10410620 | 0.13 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr4_+_11464255 | 0.12 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr8_-_43997538 | 0.10 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr12_-_19007834 | 0.10 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chrM_+_9735 | 0.10 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr16_-_22930925 | 0.08 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr16_-_21038015 | 0.08 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr14_+_23717165 | 0.07 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr22_+_4513607 | 0.07 |
ENSDART00000185696
|
ctxn1
|
cortexin 1 |
| chr4_-_75681004 | 0.07 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr3_+_12816362 | 0.07 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr21_-_43949208 | 0.06 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr24_-_25004553 | 0.06 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr15_-_37835638 | 0.06 |
ENSDART00000128922
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr10_-_34871737 | 0.04 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr17_+_33311784 | 0.03 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr1_+_37752171 | 0.03 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr19_-_32914227 | 0.03 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr7_+_52154215 | 0.03 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr14_+_25817628 | 0.02 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr21_-_21781158 | 0.02 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr21_-_15046065 | 0.02 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr19_+_31904836 | 0.00 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 1.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.3 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.2 | 1.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 1.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.6 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 1.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.0 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 1.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.8 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 5.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.9 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 2.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |