Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
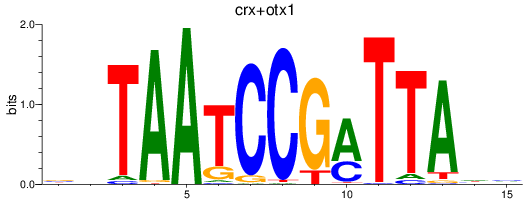
Results for crx+otx1
Z-value: 2.57
Transcription factors associated with crx+otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crx
|
ENSDARG00000011989 | cone-rod homeobox |
|
otx1
|
ENSDARG00000094992 | orthodenticle homeobox 1 |
|
crx
|
ENSDARG00000113850 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx1 | dr11_v1_chr17_+_24318753_24318753 | 0.93 | 2.7e-04 | Click! |
| crx | dr11_v1_chr5_+_36932718_36932718 | -0.49 | 1.8e-01 | Click! |
Activity profile of crx+otx1 motif
Sorted Z-values of crx+otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_45125537 | 5.83 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr16_-_46579936 | 5.53 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr10_-_42898220 | 5.26 |
ENSDART00000099270
|
CU326366.2
|
|
| chr11_-_1550709 | 5.08 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr2_-_31686353 | 4.66 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr9_-_8314028 | 4.58 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr19_+_42898239 | 3.38 |
ENSDART00000051724
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr13_+_15581270 | 3.13 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr20_-_54014539 | 3.07 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr16_-_25741225 | 2.98 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr22_+_1170294 | 2.92 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr20_-_54014373 | 2.84 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr22_+_25453334 | 2.83 |
ENSDART00000123962
|
si:ch211-12h2.6
|
si:ch211-12h2.6 |
| chr20_+_1272526 | 2.81 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr23_+_2560005 | 2.70 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr3_-_3366590 | 2.67 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr13_+_1015749 | 2.67 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr14_+_30340251 | 2.65 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr16_-_16225260 | 2.57 |
ENSDART00000165790
|
gra
|
granulito |
| chr20_+_18260358 | 2.57 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr20_+_42537768 | 2.55 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr23_+_7710721 | 2.48 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr24_+_10039165 | 2.47 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr5_-_69180587 | 2.45 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr2_-_39036604 | 2.44 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr1_+_52633367 | 2.36 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr10_+_6013076 | 2.36 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr8_+_25091407 | 2.35 |
ENSDART00000143922
|
atxn7l2b
|
ataxin 7-like 2b |
| chr7_+_39011852 | 2.23 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_+_25683069 | 2.22 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr8_+_2656681 | 2.21 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr5_-_23715027 | 2.18 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_-_15679107 | 2.15 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr5_-_69180227 | 2.14 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr15_-_5580093 | 2.11 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr1_-_24349759 | 2.09 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr4_+_68562464 | 2.06 |
ENSDART00000192954
|
BX548011.4
|
|
| chr17_+_50701748 | 2.06 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr4_-_17353100 | 2.04 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr8_+_28724692 | 2.02 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr7_-_39378903 | 2.02 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr23_+_39963599 | 2.00 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr10_+_42898103 | 2.00 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr8_+_2656231 | 1.97 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr20_+_2134816 | 1.96 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_-_27994679 | 1.94 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr18_+_27439680 | 1.92 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr14_-_6402769 | 1.90 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr5_-_27993972 | 1.88 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr5_-_23715861 | 1.86 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr12_+_23850661 | 1.85 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr5_-_52216170 | 1.82 |
ENSDART00000158542
ENSDART00000192981 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr13_+_8840772 | 1.80 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr2_+_44972720 | 1.77 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr16_+_53519048 | 1.72 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr21_-_18262287 | 1.70 |
ENSDART00000176716
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr18_+_3634652 | 1.68 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr7_+_39011355 | 1.67 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr25_-_10610961 | 1.66 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr24_-_31140356 | 1.66 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr25_+_35891342 | 1.65 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr6_+_37655078 | 1.64 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr10_+_39476600 | 1.58 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr9_-_2945008 | 1.57 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr7_+_20030888 | 1.53 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr1_+_44523516 | 1.52 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr9_-_5318873 | 1.52 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr11_+_6295370 | 1.51 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr9_-_135774 | 1.50 |
ENSDART00000160435
|
FQ377903.1
|
|
| chr18_+_173603 | 1.49 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr9_+_17982737 | 1.48 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr17_-_1705013 | 1.47 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr9_-_46276626 | 1.45 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr20_-_36617313 | 1.45 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr16_+_9726038 | 1.43 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr17_+_18031304 | 1.41 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr1_-_43712120 | 1.40 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr2_-_32387441 | 1.38 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr10_-_35108683 | 1.38 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr14_+_21754521 | 1.37 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr2_-_42864472 | 1.37 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr20_-_2134620 | 1.36 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr11_-_27821 | 1.35 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr18_-_8030073 | 1.34 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr23_+_25893020 | 1.31 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr23_-_42810664 | 1.28 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr18_-_19006634 | 1.27 |
ENSDART00000060731
|
ints14
|
integrator complex subunit 14 |
| chr4_-_73562122 | 1.27 |
ENSDART00000174096
|
CU570689.2
|
|
| chr4_-_28353538 | 1.26 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr15_-_26930999 | 1.26 |
ENSDART00000181674
ENSDART00000126046 |
ccdc9
|
coiled-coil domain containing 9 |
| chr18_-_19005919 | 1.26 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr2_-_37537887 | 1.25 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr22_-_16154771 | 1.25 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr4_-_11053543 | 1.24 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr22_-_12304591 | 1.23 |
ENSDART00000136408
|
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr19_-_12078583 | 1.20 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr16_+_38337783 | 1.19 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr11_-_25257595 | 1.18 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr25_-_32363341 | 1.18 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr4_+_72797711 | 1.15 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr20_+_54666222 | 1.15 |
ENSDART00000166592
|
CABZ01087948.1
|
|
| chr7_+_22330466 | 1.14 |
ENSDART00000187347
ENSDART00000174483 |
fgf11a
|
fibroblast growth factor 11a |
| chr23_+_21492151 | 1.13 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr12_-_33659328 | 1.12 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr1_-_45580787 | 1.11 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_+_15069011 | 1.11 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr2_+_24080694 | 1.10 |
ENSDART00000024058
|
kcnh2a
|
potassium voltage-gated channel, subfamily H (eag-related), member 2a |
| chr21_-_40562705 | 1.08 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr6_+_37623693 | 1.08 |
ENSDART00000144812
ENSDART00000182709 |
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr4_+_5196469 | 1.07 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr16_+_19637384 | 1.06 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr23_+_12361899 | 1.00 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr5_+_11407504 | 0.99 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr15_+_20355465 | 0.98 |
ENSDART00000143092
ENSDART00000062666 |
il15l
|
interleukin 15, like |
| chr8_-_51806584 | 0.98 |
ENSDART00000146976
|
si:ch73-313l22.2
|
si:ch73-313l22.2 |
| chr21_-_2209012 | 0.97 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr3_-_26183699 | 0.96 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_-_42876596 | 0.95 |
ENSDART00000086156
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr25_-_10630496 | 0.94 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr3_-_15210491 | 0.93 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr3_-_13068189 | 0.93 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr18_+_5618368 | 0.92 |
ENSDART00000159945
|
ulk3
|
unc-51 like kinase 3 |
| chr9_+_3519191 | 0.91 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr22_+_30047245 | 0.91 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr11_+_19080400 | 0.89 |
ENSDART00000044423
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr18_-_10713414 | 0.88 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr2_+_36112273 | 0.88 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr5_-_32489796 | 0.87 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr4_-_73561848 | 0.87 |
ENSDART00000174210
|
CU570689.2
|
|
| chr8_+_18545933 | 0.84 |
ENSDART00000148806
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr5_-_22868650 | 0.84 |
ENSDART00000057541
|
apool
|
apolipoprotein O-like |
| chr15_-_26931541 | 0.83 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr3_+_62000822 | 0.79 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr25_+_7532627 | 0.78 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr18_+_10784730 | 0.78 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr4_+_43408004 | 0.77 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr17_+_10593398 | 0.76 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr25_-_19090479 | 0.76 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr14_+_11289851 | 0.75 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr6_-_15641686 | 0.75 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr3_-_26184018 | 0.73 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr25_+_29694158 | 0.72 |
ENSDART00000174568
|
brd1b
|
bromodomain containing 1b |
| chr22_+_2431585 | 0.69 |
ENSDART00000167758
|
zgc:171435
|
zgc:171435 |
| chr1_+_59328030 | 0.68 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr17_+_18031899 | 0.67 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr13_+_2442841 | 0.66 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr7_-_30492261 | 0.64 |
ENSDART00000173954
|
adam10a
|
ADAM metallopeptidase domain 10a |
| chr3_-_20058417 | 0.62 |
ENSDART00000165695
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr13_-_9101236 | 0.62 |
ENSDART00000163266
|
HTRA2 (1 of many)
|
si:dkey-19p15.4 |
| chr22_-_7457247 | 0.62 |
ENSDART00000106081
|
BX511034.2
|
|
| chr15_+_20364595 | 0.60 |
ENSDART00000062676
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr2_-_15349382 | 0.59 |
ENSDART00000057238
|
olfm3b
|
olfactomedin 3b |
| chr9_+_1313418 | 0.59 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr16_+_38159758 | 0.58 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr3_+_1209006 | 0.58 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr1_-_45215343 | 0.57 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr4_+_31450683 | 0.56 |
ENSDART00000169303
|
znf996
|
zinc finger protein 996 |
| chr17_+_2130018 | 0.56 |
ENSDART00000193675
ENSDART00000110529 |
bub1bb
|
BUB1 mitotic checkpoint serine/threonine kinase Bb |
| chr10_+_39476432 | 0.55 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr25_+_32031536 | 0.55 |
ENSDART00000173656
|
tjp1b
|
tight junction protein 1b |
| chr3_+_22327738 | 0.54 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr25_-_13549577 | 0.53 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr21_+_33503835 | 0.52 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr8_+_53120278 | 0.52 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr16_+_33987892 | 0.50 |
ENSDART00000166302
|
pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr15_+_20363859 | 0.49 |
ENSDART00000166846
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr22_-_38934989 | 0.49 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr15_-_18213515 | 0.48 |
ENSDART00000101635
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr11_-_15090118 | 0.45 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr7_-_30492018 | 0.45 |
ENSDART00000099639
ENSDART00000162705 ENSDART00000173663 |
adam10a
|
ADAM metallopeptidase domain 10a |
| chr9_+_1365747 | 0.44 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr14_+_2487672 | 0.44 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr8_-_46572298 | 0.41 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr3_+_32842825 | 0.41 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr4_-_20485508 | 0.40 |
ENSDART00000183276
ENSDART00000023337 |
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr17_+_30448452 | 0.40 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr4_+_36906436 | 0.39 |
ENSDART00000136773
|
BX537137.1
|
|
| chr3_+_50602866 | 0.38 |
ENSDART00000190069
ENSDART00000153921 |
gsg1l2a
|
gsg1-like 2a |
| chr11_+_29770966 | 0.37 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr12_-_35883814 | 0.37 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr10_+_26652859 | 0.36 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr14_+_52413846 | 0.35 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr5_-_65082107 | 0.35 |
ENSDART00000162511
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr25_-_10629940 | 0.33 |
ENSDART00000154483
ENSDART00000155231 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr25_+_35913614 | 0.33 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr13_+_41917606 | 0.33 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr19_-_2876321 | 0.33 |
ENSDART00000159253
|
exosc7
|
exosome component 7 |
| chr23_-_35064785 | 0.33 |
ENSDART00000172240
|
BX294434.1
|
|
| chr20_-_54924593 | 0.31 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr14_-_8816933 | 0.30 |
ENSDART00000163383
|
si:dkeyp-115e12.7
|
si:dkeyp-115e12.7 |
| chr5_+_26847190 | 0.30 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr14_-_36412473 | 0.29 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr2_+_24722771 | 0.28 |
ENSDART00000147207
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr19_+_34169055 | 0.27 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr23_+_30827959 | 0.27 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr4_+_65281752 | 0.27 |
ENSDART00000183934
|
znf1027
|
zinc finger protein 1027 |
| chr13_-_40499296 | 0.26 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr6_-_39218609 | 0.26 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr17_-_7371564 | 0.25 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr23_+_1181248 | 0.24 |
ENSDART00000170942
|
utrn
|
utrophin |
Network of associatons between targets according to the STRING database.
First level regulatory network of crx+otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.5 | 1.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 2.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 1.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 2.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 1.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 2.5 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 2.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 2.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 4.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 1.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 2.7 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 1.1 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 | 0.7 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 0.5 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 2.7 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 1.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 3.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 0.5 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 1.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 4.9 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 1.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 2.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.4 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 1.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 2.9 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 1.1 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 2.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 2.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 2.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.8 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 2.2 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 2.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 2.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 2.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.8 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.6 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.7 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.5 | GO:0001756 | somitogenesis(GO:0001756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 2.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 1.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 2.6 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.5 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 4.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.5 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.6 | 3.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 1.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 2.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 1.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.3 | 2.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.9 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.2 | 3.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 1.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 2.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 5.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 8.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.2 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 2.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 3.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 7.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 2.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 6.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 2.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |