Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
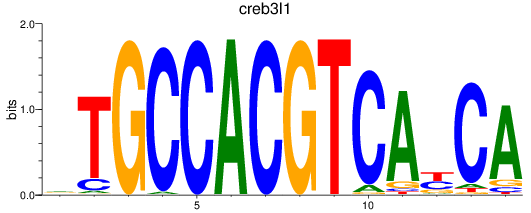
Results for creb3l1
Z-value: 0.88
Transcription factors associated with creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l1
|
ENSDARG00000015793 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l1 | dr11_v1_chr7_+_38898208_38898208 | -0.91 | 6.7e-04 | Click! |
Activity profile of creb3l1 motif
Sorted Z-values of creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43636595 | 1.97 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr12_+_30789611 | 1.60 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr9_+_45428041 | 1.55 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr17_+_10090638 | 1.54 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr19_+_33139164 | 1.38 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr4_-_26095755 | 1.26 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr18_+_2153530 | 1.25 |
ENSDART00000158255
|
pygo1
|
pygopus family PHD finger 1 |
| chr12_-_35582683 | 1.21 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr21_+_10701834 | 1.16 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr18_-_7481036 | 1.16 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr4_+_68562464 | 1.14 |
ENSDART00000192954
|
BX548011.4
|
|
| chr21_+_10702031 | 1.11 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr6_-_50685862 | 1.11 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr12_-_35582521 | 1.09 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr18_-_7031409 | 1.09 |
ENSDART00000148485
ENSDART00000005405 |
calub
|
calumenin b |
| chr5_-_48680580 | 1.08 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr9_+_34230067 | 1.04 |
ENSDART00000139157
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr9_+_20554178 | 1.02 |
ENSDART00000183911
ENSDART00000147435 |
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_+_54124209 | 0.98 |
ENSDART00000187730
|
LO017722.1
|
|
| chr20_+_22799857 | 0.95 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr9_+_20554896 | 0.91 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr10_+_15603082 | 0.82 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr5_-_28968964 | 0.81 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr14_+_28281744 | 0.81 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr12_+_13348918 | 0.80 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr22_-_21897203 | 0.76 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr11_-_36957127 | 0.74 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr8_+_47219107 | 0.71 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr3_+_32862730 | 0.69 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr3_-_26183699 | 0.66 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr1_-_18592068 | 0.64 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr7_+_13491452 | 0.63 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr23_-_306796 | 0.63 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr3_-_26184018 | 0.62 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr15_-_18115540 | 0.61 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr5_+_61556172 | 0.60 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr14_-_45595711 | 0.59 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr15_+_32249062 | 0.58 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr20_+_22799641 | 0.57 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr7_+_30988570 | 0.57 |
ENSDART00000180613
ENSDART00000185625 |
tjp1a
|
tight junction protein 1a |
| chr19_+_7549854 | 0.54 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr22_+_24157807 | 0.54 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_-_36819624 | 0.49 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr20_-_1191910 | 0.49 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr16_+_5612547 | 0.45 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr23_-_25798099 | 0.43 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr4_-_5795309 | 0.40 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr6_-_59388625 | 0.40 |
ENSDART00000111992
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr17_-_11417551 | 0.39 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr6_-_9236309 | 0.39 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr11_-_2456656 | 0.38 |
ENSDART00000171219
ENSDART00000167938 |
slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr6_-_13716744 | 0.35 |
ENSDART00000111102
|
chpfb
|
chondroitin polymerizing factor b |
| chr9_+_19529951 | 0.33 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr14_-_45464112 | 0.33 |
ENSDART00000173209
|
ehd1a
|
EH-domain containing 1a |
| chr19_+_20708460 | 0.30 |
ENSDART00000034124
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr24_+_854877 | 0.30 |
ENSDART00000188408
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr17_+_30448452 | 0.28 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr16_+_19637384 | 0.28 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr5_-_30516646 | 0.25 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr21_+_5932140 | 0.22 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr18_-_18587745 | 0.20 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr22_-_775776 | 0.18 |
ENSDART00000149749
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr5_+_58996765 | 0.17 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr14_-_26411918 | 0.17 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr22_+_31295791 | 0.16 |
ENSDART00000092447
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr14_+_8594367 | 0.16 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr18_-_3542435 | 0.16 |
ENSDART00000184017
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr1_+_277731 | 0.15 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr18_+_13792490 | 0.14 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr13_-_40499296 | 0.12 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr9_-_29497916 | 0.11 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr13_-_9875538 | 0.11 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr4_-_2637689 | 0.10 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr18_+_7553950 | 0.05 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr4_-_17812131 | 0.04 |
ENSDART00000025731
|
spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr10_-_3427589 | 0.03 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr25_-_8513255 | 0.03 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr2_+_5948534 | 0.01 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr7_+_18100996 | 0.00 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 0.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.6 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 1.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.7 | GO:0009086 | methionine biosynthetic process(GO:0009086) tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 2.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.5 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.9 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 2.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 4.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 4.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |