Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
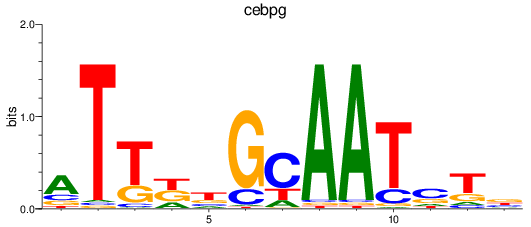
Results for cebpg
Z-value: 0.23
Transcription factors associated with cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpg
|
ENSDARG00000036073 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpg | dr11_v1_chr7_+_38090515_38090515 | -0.63 | 6.8e-02 | Click! |
Activity profile of cebpg motif
Sorted Z-values of cebpg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_46818001 | 0.28 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr22_-_23668356 | 0.27 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr9_-_9671244 | 0.26 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr16_+_23984179 | 0.25 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr1_-_10071422 | 0.23 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr10_-_36808348 | 0.23 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr12_+_25775734 | 0.23 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr25_+_24616717 | 0.22 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr24_-_32665283 | 0.22 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr9_+_9502610 | 0.22 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr3_-_57666518 | 0.21 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr25_-_3830272 | 0.21 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr8_-_50147948 | 0.20 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr5_-_63509581 | 0.19 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr15_-_21014270 | 0.19 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr3_+_23703704 | 0.18 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr12_-_35949936 | 0.18 |
ENSDART00000192583
|
AL954682.1
|
|
| chr10_-_17103651 | 0.17 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr20_-_30900947 | 0.16 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr8_-_4475908 | 0.16 |
ENSDART00000191027
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr2_+_4207209 | 0.16 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr23_-_33986762 | 0.16 |
ENSDART00000144609
|
tmed4
|
transmembrane p24 trafficking protein 4 |
| chr7_+_56577906 | 0.15 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr10_-_22803740 | 0.15 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr4_+_17279966 | 0.14 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr5_+_39504136 | 0.14 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr13_+_30903816 | 0.14 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr21_-_4539899 | 0.13 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr9_+_32301456 | 0.13 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr9_+_32301017 | 0.13 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr17_-_10838434 | 0.13 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr24_-_33284945 | 0.13 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr2_-_30182353 | 0.13 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr3_+_37197686 | 0.13 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr2_+_37971353 | 0.13 |
ENSDART00000165085
|
apol1
|
apolipoprotein L, 1 |
| chr6_-_345503 | 0.13 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr16_-_43356018 | 0.12 |
ENSDART00000181683
|
FO704821.1
|
|
| chr22_-_11729350 | 0.12 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr3_-_38915998 | 0.12 |
ENSDART00000141886
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr18_-_40905901 | 0.12 |
ENSDART00000064848
|
pglyrp5
|
peptidoglycan recognition protein 5 |
| chr16_+_48753664 | 0.12 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr10_-_13772847 | 0.12 |
ENSDART00000145103
ENSDART00000184491 |
cntfr
|
ciliary neurotrophic factor receptor |
| chr22_-_26236188 | 0.12 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr15_+_9327252 | 0.12 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr10_-_45002196 | 0.11 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr4_-_20292821 | 0.11 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr19_-_44054930 | 0.11 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr17_-_23412705 | 0.11 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr11_-_31172276 | 0.11 |
ENSDART00000171520
|
si:dkey-238i5.3
|
si:dkey-238i5.3 |
| chr15_-_29348212 | 0.11 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr23_+_27675581 | 0.11 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr9_+_219124 | 0.10 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr19_-_21832441 | 0.10 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr24_-_25184553 | 0.10 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_+_53482597 | 0.10 |
ENSDART00000180333
|
BX323994.1
|
|
| chr22_-_26175237 | 0.10 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr3_+_40809011 | 0.10 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr6_-_609880 | 0.09 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr5_-_58840971 | 0.09 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr15_+_7064819 | 0.09 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr9_-_4856767 | 0.09 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr1_+_14253118 | 0.09 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr10_+_21776911 | 0.09 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr20_-_1151265 | 0.09 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr3_+_54581987 | 0.09 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr16_-_42004544 | 0.09 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr10_-_41980797 | 0.09 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr6_+_27452289 | 0.09 |
ENSDART00000186265
|
sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr25_+_20715950 | 0.08 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr25_+_20716176 | 0.08 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr21_-_40174647 | 0.08 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr8_+_1082100 | 0.08 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr22_-_10487490 | 0.08 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr3_-_38745598 | 0.08 |
ENSDART00000127483
|
si:ch211-198m17.1
|
si:ch211-198m17.1 |
| chr10_+_15310811 | 0.08 |
ENSDART00000136239
|
kcnv2a
|
potassium channel, subfamily V, member 2a |
| chr21_-_45077429 | 0.08 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr23_-_4915118 | 0.08 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr3_-_22216442 | 0.08 |
ENSDART00000153613
|
maptb
|
microtubule-associated protein tau b |
| chr21_+_21309164 | 0.08 |
ENSDART00000132174
|
si:ch211-191j22.8
|
si:ch211-191j22.8 |
| chr2_-_40199780 | 0.07 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr4_+_42604252 | 0.07 |
ENSDART00000184850
|
CR925768.1
|
|
| chr18_-_5509616 | 0.07 |
ENSDART00000142945
|
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr23_-_24195519 | 0.07 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr5_+_45677781 | 0.07 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr5_-_24238733 | 0.07 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr3_-_16055432 | 0.07 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr6_+_13779857 | 0.07 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr24_+_38671054 | 0.07 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr24_+_16905188 | 0.07 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr23_-_16843649 | 0.07 |
ENSDART00000129971
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr4_-_5595237 | 0.07 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr12_-_21834611 | 0.07 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr13_+_29193649 | 0.06 |
ENSDART00000045951
|
unc5b
|
unc-5 netrin receptor B |
| chr5_+_54400971 | 0.06 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr14_-_38929885 | 0.06 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr15_+_28355023 | 0.06 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr22_+_7486867 | 0.06 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr15_-_22139566 | 0.06 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr8_-_53488832 | 0.06 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr2_-_44746723 | 0.06 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr4_+_30776883 | 0.06 |
ENSDART00000169519
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr5_+_17727310 | 0.06 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr11_+_43419809 | 0.06 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr18_-_16179129 | 0.06 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr1_+_17892944 | 0.06 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr4_-_17793152 | 0.06 |
ENSDART00000134080
|
mybpc1
|
myosin binding protein C, slow type |
| chr24_+_13925066 | 0.06 |
ENSDART00000134221
ENSDART00000012253 ENSDART00000081595 ENSDART00000136443 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_-_23575007 | 0.05 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr20_-_40755614 | 0.05 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr25_-_29415369 | 0.05 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr4_-_37479395 | 0.05 |
ENSDART00000168299
ENSDART00000157864 |
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr15_-_3282220 | 0.05 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr19_+_16015881 | 0.05 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr6_-_39344259 | 0.05 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr1_+_58067815 | 0.04 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr17_-_14613711 | 0.04 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr25_-_3469576 | 0.04 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr7_+_49862837 | 0.04 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr1_+_45056371 | 0.04 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr10_+_15088534 | 0.04 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr7_+_23580082 | 0.04 |
ENSDART00000183245
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr9_+_38292947 | 0.04 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr24_-_26485098 | 0.04 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr8_-_36140405 | 0.04 |
ENSDART00000182806
|
CT583723.2
|
|
| chr2_+_30948503 | 0.04 |
ENSDART00000149923
|
myom1a
|
myomesin 1a (skelemin) |
| chr18_+_34225520 | 0.04 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr20_-_32186886 | 0.04 |
ENSDART00000170279
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr9_-_21838045 | 0.04 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr20_+_32118559 | 0.04 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr23_-_1658980 | 0.04 |
ENSDART00000186227
|
CU693481.1
|
|
| chr23_+_21405201 | 0.04 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr6_+_13117598 | 0.04 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr11_-_44163164 | 0.04 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr7_+_65533307 | 0.04 |
ENSDART00000156301
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr16_+_28601974 | 0.04 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr10_-_20357013 | 0.04 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr12_-_22524388 | 0.04 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr22_-_18491813 | 0.03 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr4_+_37936249 | 0.03 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr17_+_12058509 | 0.03 |
ENSDART00000150209
|
tfb2m
|
transcription factor B2, mitochondrial |
| chr19_+_41080240 | 0.03 |
ENSDART00000087295
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr23_-_40814080 | 0.03 |
ENSDART00000135713
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr23_-_40792128 | 0.03 |
ENSDART00000145360
|
si:dkey-194e6.2
|
si:dkey-194e6.2 |
| chr15_+_31303355 | 0.03 |
ENSDART00000143423
|
or116-2
|
odorant receptor, family F, subfamily 116, member 2 |
| chr19_-_9662958 | 0.03 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr22_-_13544244 | 0.03 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr10_-_7785930 | 0.03 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr7_-_69121896 | 0.03 |
ENSDART00000130227
|
crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr14_+_7377552 | 0.03 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr23_-_24195723 | 0.03 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr21_-_22709251 | 0.03 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr7_-_58289847 | 0.02 |
ENSDART00000012822
|
CR354540.1
|
|
| chr23_-_11130683 | 0.02 |
ENSDART00000181189
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr25_+_34811070 | 0.02 |
ENSDART00000087410
|
adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr11_+_25044082 | 0.02 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr17_-_45155181 | 0.02 |
ENSDART00000185274
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr9_-_33785093 | 0.02 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr12_-_9087566 | 0.02 |
ENSDART00000152793
|
exoc6
|
exocyst complex component 6 |
| chr25_-_3087556 | 0.02 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr19_-_22507715 | 0.02 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr3_+_22242269 | 0.02 |
ENSDART00000168970
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr8_-_18582922 | 0.02 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr20_+_23440632 | 0.02 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr12_+_46512881 | 0.02 |
ENSDART00000105454
|
CU855711.1
|
|
| chr23_-_36003441 | 0.02 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr25_-_16076257 | 0.02 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr10_+_6496185 | 0.02 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr5_+_41510387 | 0.02 |
ENSDART00000023779
|
vcp
|
valosin containing protein |
| chr5_-_29488245 | 0.02 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr10_-_28193642 | 0.02 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr22_-_10570749 | 0.01 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr2_+_31476065 | 0.01 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr17_-_23603263 | 0.01 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr20_+_40457599 | 0.01 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr4_+_9508505 | 0.01 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr21_-_43949208 | 0.01 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr4_-_9549693 | 0.01 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr7_+_33152723 | 0.01 |
ENSDART00000132658
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr1_+_30551777 | 0.01 |
ENSDART00000112778
|
gpr183b
|
G protein-coupled receptor 183b |
| chr17_+_9040165 | 0.01 |
ENSDART00000181221
ENSDART00000181846 |
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr21_+_4540127 | 0.01 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr1_+_31674297 | 0.01 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr25_-_19443421 | 0.01 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr18_+_17827149 | 0.01 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr21_+_25057176 | 0.01 |
ENSDART00000183975
|
zgc:171740
|
zgc:171740 |
| chr3_-_23574622 | 0.01 |
ENSDART00000176012
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr8_+_22955478 | 0.01 |
ENSDART00000099911
|
zgc:136605
|
zgc:136605 |
| chr2_+_11029138 | 0.00 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr4_-_73049281 | 0.00 |
ENSDART00000169165
|
LO018021.1
|
|
| chr19_+_7759354 | 0.00 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr13_-_31878043 | 0.00 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr5_-_50638905 | 0.00 |
ENSDART00000180842
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr12_+_46483618 | 0.00 |
ENSDART00000186970
|
BX005305.5
|
|
| chr25_-_16146851 | 0.00 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr2_+_42247560 | 0.00 |
ENSDART00000101230
ENSDART00000143094 |
ftr06
|
finTRIM family, member 6 |
| chr12_+_46443477 | 0.00 |
ENSDART00000191873
|
BX005305.1
|
|
| chr12_+_46425800 | 0.00 |
ENSDART00000191965
|
BX005305.1
|
|
| chr18_+_20869923 | 0.00 |
ENSDART00000138471
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr2_-_31537521 | 0.00 |
ENSDART00000148244
|
mrc1b
|
mannose receptor, C type 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0034138 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |