Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
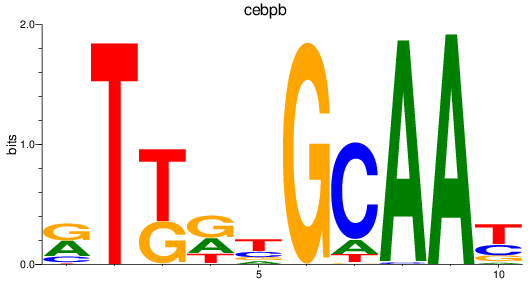
Results for cebpb
Z-value: 1.96
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr11_v1_chr8_-_28449782_28449782 | 0.26 | 5.0e-01 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50147948 | 4.61 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr14_+_36885524 | 4.05 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr16_-_21785261 | 3.84 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr5_-_63509581 | 3.71 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr5_-_30615901 | 3.56 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr12_-_5120175 | 3.52 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr3_+_30921246 | 3.09 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr15_+_32711663 | 3.09 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr9_+_3388099 | 3.07 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr14_-_12071679 | 2.80 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr3_-_46818001 | 2.80 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr2_-_40199780 | 2.75 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr14_-_12071447 | 2.68 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr11_-_24681292 | 2.61 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr20_+_16743056 | 2.61 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr24_-_32665283 | 2.33 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr3_+_40809011 | 2.31 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr23_+_44732863 | 2.26 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr9_-_1702648 | 2.24 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr7_-_60831082 | 2.20 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr5_+_39504136 | 2.16 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr20_+_10538025 | 2.14 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr16_-_16701718 | 2.12 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr6_+_21202639 | 2.12 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr20_+_10544100 | 2.06 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_9472893 | 2.06 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr1_+_26356360 | 2.03 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr17_+_52822831 | 2.02 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr3_-_49566364 | 2.00 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr1_-_10071422 | 2.00 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr16_+_23984179 | 1.97 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr3_+_23703704 | 1.97 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr25_+_31323978 | 1.96 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr17_+_10318071 | 1.92 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr10_-_17103651 | 1.91 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr23_+_2669 | 1.91 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr13_+_39532050 | 1.91 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr12_-_10409961 | 1.90 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr17_+_30587333 | 1.89 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr23_+_36095260 | 1.85 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr15_+_29085955 | 1.84 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr6_-_43449013 | 1.83 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr10_+_38610741 | 1.82 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr11_-_6265574 | 1.79 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr22_-_17595310 | 1.79 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr25_+_13191391 | 1.78 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr5_-_20195350 | 1.70 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr7_-_66452792 | 1.68 |
ENSDART00000170701
ENSDART00000082604 |
galnt18b
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18b |
| chr23_+_22656477 | 1.68 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr24_-_33284945 | 1.67 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr9_+_219124 | 1.67 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr3_-_12930217 | 1.64 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr24_-_17049270 | 1.64 |
ENSDART00000175508
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr3_+_31933893 | 1.63 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr25_+_13191615 | 1.62 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr21_-_4032650 | 1.60 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr9_-_34915351 | 1.60 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr5_+_26795773 | 1.59 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr9_-_1978090 | 1.58 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr5_+_43530388 | 1.57 |
ENSDART00000190254
ENSDART00000097618 ENSDART00000133006 |
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr1_+_26356205 | 1.57 |
ENSDART00000190064
ENSDART00000176380 |
tet2
|
tet methylcytosine dioxygenase 2 |
| chr9_+_32301017 | 1.56 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr11_+_43419809 | 1.55 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr13_+_23157053 | 1.55 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr6_-_345503 | 1.53 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr9_+_23665777 | 1.52 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr16_+_20895904 | 1.52 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr14_+_21106444 | 1.51 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr14_-_30704075 | 1.47 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr1_-_22412042 | 1.43 |
ENSDART00000074678
|
chrnb3a
|
cholinergic receptor, nicotinic, beta polypeptide 3a |
| chr12_-_31012741 | 1.42 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr21_-_15929041 | 1.42 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr9_-_9989660 | 1.41 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr6_+_58543336 | 1.40 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr23_-_36724575 | 1.39 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr19_-_5699703 | 1.39 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr8_-_23684659 | 1.35 |
ENSDART00000136865
|
cfp
|
complement factor properdin |
| chr19_+_4990320 | 1.35 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr6_-_11768198 | 1.35 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr8_+_25351863 | 1.35 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr19_-_9712530 | 1.32 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr5_-_55395384 | 1.31 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr17_-_43466317 | 1.31 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr20_-_26531850 | 1.30 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr21_-_36972127 | 1.28 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr14_-_1098607 | 1.28 |
ENSDART00000169090
ENSDART00000158905 |
FQ311908.1
|
|
| chr12_+_31616412 | 1.28 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr7_+_63325819 | 1.27 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr11_+_23933016 | 1.27 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr14_+_21107032 | 1.26 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr21_-_39058490 | 1.26 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr6_+_54142311 | 1.25 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr7_+_11543999 | 1.24 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr22_-_26236188 | 1.23 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr22_-_26175237 | 1.22 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr7_-_4296771 | 1.22 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr22_+_17203752 | 1.21 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr20_-_26532167 | 1.20 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr2_-_32352946 | 1.20 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr1_-_59130695 | 1.19 |
ENSDART00000152560
|
FP015850.1
|
|
| chr5_-_33259079 | 1.19 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr7_+_33314925 | 1.17 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr10_-_36808348 | 1.17 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr12_+_25775734 | 1.17 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr15_-_17960228 | 1.16 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr5_-_58840971 | 1.16 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr16_+_50289916 | 1.14 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr17_+_52823015 | 1.14 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr10_+_21807497 | 1.14 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr21_-_40174647 | 1.13 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr8_+_31119548 | 1.13 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr4_-_12914163 | 1.12 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr6_-_18960105 | 1.11 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr25_-_3469576 | 1.10 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr22_-_10487490 | 1.10 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr23_+_11285662 | 1.09 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr10_+_17088261 | 1.07 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr25_-_2081371 | 1.07 |
ENSDART00000104915
ENSDART00000156925 |
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr25_+_35553542 | 1.06 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr9_+_907459 | 1.04 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr24_-_36727922 | 1.02 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr1_-_59126139 | 1.01 |
ENSDART00000156105
|
si:ch1073-110a20.7
|
si:ch1073-110a20.7 |
| chr19_-_9829965 | 1.00 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr12_-_17199381 | 1.00 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr10_+_38593645 | 0.99 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr22_-_18491813 | 0.99 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr15_+_28355023 | 0.99 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr9_+_32301456 | 0.99 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr15_-_29573267 | 0.98 |
ENSDART00000099947
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr19_+_5674907 | 0.98 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr12_+_35119762 | 0.98 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr7_-_52963493 | 0.97 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr1_+_14253118 | 0.97 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr5_-_45894802 | 0.96 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr9_+_9502610 | 0.95 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr17_+_443264 | 0.95 |
ENSDART00000159086
|
zgc:195050
|
zgc:195050 |
| chr7_-_7845540 | 0.94 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr3_-_57744323 | 0.92 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr14_-_17072736 | 0.92 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr19_+_1370504 | 0.91 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr5_-_43935460 | 0.91 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr21_-_5007109 | 0.90 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr18_-_20869175 | 0.90 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr19_+_4990496 | 0.90 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr1_+_50538839 | 0.90 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr25_-_27843066 | 0.90 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr9_+_13985567 | 0.89 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr1_-_59130383 | 0.88 |
ENSDART00000171552
|
FP015850.1
|
|
| chr19_-_33212023 | 0.87 |
ENSDART00000189209
|
trib1
|
tribbles pseudokinase 1 |
| chr9_+_38292947 | 0.86 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr2_-_30460293 | 0.86 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr19_+_8606883 | 0.85 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr4_-_12725513 | 0.85 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr8_-_2434282 | 0.84 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr5_-_39474235 | 0.84 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr12_-_10038870 | 0.83 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr10_+_22771176 | 0.82 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr6_+_16406723 | 0.82 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr4_-_5595237 | 0.81 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr1_-_59134522 | 0.80 |
ENSDART00000152764
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr19_-_34927201 | 0.80 |
ENSDART00000076518
|
sla1
|
Src-like-adaptor 1 |
| chr5_-_69437422 | 0.80 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr9_-_7238839 | 0.80 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr15_-_22139566 | 0.80 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr1_-_59104145 | 0.79 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr24_+_119680 | 0.77 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr5_-_43935119 | 0.77 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr22_+_10201826 | 0.76 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr25_+_19485198 | 0.76 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr4_+_12617108 | 0.75 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr25_-_17239008 | 0.75 |
ENSDART00000151965
ENSDART00000152106 ENSDART00000152107 |
ccnd2a
|
cyclin D2, a |
| chr19_-_44054930 | 0.73 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr5_+_32162684 | 0.73 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr8_-_28349859 | 0.73 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr25_+_7784582 | 0.72 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr1_-_59141715 | 0.71 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr7_-_22941472 | 0.70 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr25_+_31222069 | 0.69 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr6_-_32025225 | 0.69 |
ENSDART00000006417
|
pgm1
|
phosphoglucomutase 1 |
| chr7_+_4162994 | 0.67 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr6_+_27151940 | 0.67 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr21_-_30111134 | 0.66 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr12_-_14922955 | 0.66 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr18_+_507835 | 0.66 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr18_-_16179129 | 0.66 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr20_+_23440632 | 0.65 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr7_-_35036770 | 0.65 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr2_-_3044947 | 0.64 |
ENSDART00000192642
|
guk1a
|
guanylate kinase 1a |
| chr6_+_58915889 | 0.64 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr13_+_28705143 | 0.64 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr2_+_7715810 | 0.64 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr5_+_53482597 | 0.63 |
ENSDART00000180333
|
BX323994.1
|
|
| chr16_+_14029283 | 0.63 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr25_+_13498188 | 0.62 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr9_-_4856767 | 0.62 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr25_-_32311048 | 0.61 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr11_+_37251825 | 0.60 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr23_+_25305431 | 0.59 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr12_-_46959990 | 0.59 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr17_-_45552602 | 0.58 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.6 | 1.8 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.6 | 2.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.6 | 2.9 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 1.6 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.5 | 3.6 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.5 | 3.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 3.5 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.5 | 1.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.5 | 4.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 1.7 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 1.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.4 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 1.0 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.3 | 0.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 0.8 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.3 | 1.6 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.3 | 1.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 5.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 1.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.8 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.2 | 2.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 0.6 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.2 | 0.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 1.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 1.9 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.2 | 3.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.7 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 1.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 2.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 2.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 2.3 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 2.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 1.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 2.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 2.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.1 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.8 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 2.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.0 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 0.5 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.8 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.7 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 2.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.5 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 2.9 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 2.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 3.1 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.6 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 1.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.5 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.4 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 2.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 1.1 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.8 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.3 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 1.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 1.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 3.2 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.9 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 1.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.0 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 0.7 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.0 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 1.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 2.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 2.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.5 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 37.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 2.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.5 | 3.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 3.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 2.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.4 | 1.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 1.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 2.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 2.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.3 | 1.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 1.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 1.0 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 4.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.6 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 1.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 4.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 3.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 6.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 4.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 5.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 3.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |