Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
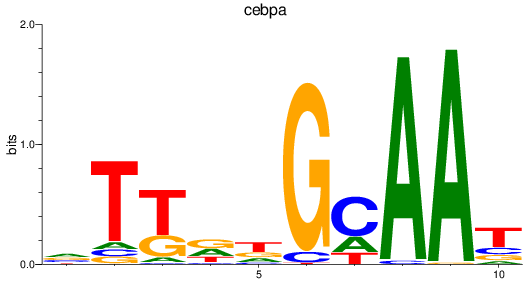
Results for cebpa
Z-value: 1.25
Transcription factors associated with cebpa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpa
|
ENSDARG00000036074 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpa | dr11_v1_chr7_-_38087865_38087865 | 0.40 | 2.9e-01 | Click! |
Activity profile of cebpa motif
Sorted Z-values of cebpa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50147948 | 3.06 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr14_+_36885524 | 2.28 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr12_+_15008582 | 1.87 |
ENSDART00000003847
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr12_+_15002757 | 1.82 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr16_-_21785261 | 1.62 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr2_-_40199780 | 1.39 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr14_+_46313396 | 1.27 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_-_24103666 | 1.26 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr5_-_30615901 | 1.25 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr15_+_32711663 | 1.21 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr15_+_28202170 | 1.17 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr2_-_5728843 | 1.15 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr12_-_16898140 | 1.13 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr5_-_63509581 | 1.04 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr8_+_13503377 | 1.03 |
ENSDART00000034740
ENSDART00000167187 |
fut9d
|
fucosyltransferase 9d |
| chr14_-_25949713 | 1.03 |
ENSDART00000181455
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr3_-_46818001 | 1.00 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_-_125091 | 0.99 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr21_-_4032650 | 0.92 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr12_-_16720196 | 0.91 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr21_-_10773344 | 0.90 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr16_+_23984179 | 0.90 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr14_-_25949951 | 0.89 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr11_-_39044595 | 0.86 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr9_+_51265283 | 0.85 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr7_-_8438657 | 0.84 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr19_-_9472893 | 0.83 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr12_-_16720432 | 0.82 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr3_-_57666518 | 0.82 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr1_-_59141715 | 0.81 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr25_+_31323978 | 0.80 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr9_+_3388099 | 0.80 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr1_+_10051763 | 0.78 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr22_-_26236188 | 0.77 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr8_-_41228530 | 0.77 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr10_+_9553935 | 0.77 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr12_-_35949936 | 0.76 |
ENSDART00000192583
|
AL954682.1
|
|
| chr5_-_14373662 | 0.75 |
ENSDART00000183694
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr17_+_10318071 | 0.75 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr6_-_49510553 | 0.73 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr10_+_38610741 | 0.71 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr25_-_32751982 | 0.70 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr3_+_31933893 | 0.70 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr6_+_21202639 | 0.69 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr1_-_10071422 | 0.69 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr5_-_20195350 | 0.69 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr3_+_40809011 | 0.69 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr9_+_32301017 | 0.68 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr9_-_1702648 | 0.66 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr8_+_22931427 | 0.66 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr6_+_2093206 | 0.65 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr1_-_59130695 | 0.63 |
ENSDART00000152560
|
FP015850.1
|
|
| chr3_-_32603191 | 0.63 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr10_-_22803740 | 0.63 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr6_+_58549080 | 0.63 |
ENSDART00000180117
|
stmn3
|
stathmin-like 3 |
| chr12_+_15666949 | 0.63 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr23_+_44732863 | 0.62 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr11_+_6116503 | 0.62 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr22_-_17595310 | 0.61 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr11_+_13629528 | 0.61 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr17_+_6276559 | 0.60 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr25_-_10564721 | 0.59 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr24_-_33284945 | 0.59 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr4_-_16345227 | 0.59 |
ENSDART00000079521
|
kera
|
keratocan |
| chr6_+_2093367 | 0.59 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr24_-_32665283 | 0.58 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr1_-_26027327 | 0.57 |
ENSDART00000171292
ENSDART00000170878 |
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr19_-_5699703 | 0.57 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr7_-_60831082 | 0.57 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr23_+_22656477 | 0.56 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr23_+_27675581 | 0.56 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr12_-_17199381 | 0.56 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr9_+_24008879 | 0.55 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr17_+_443264 | 0.55 |
ENSDART00000159086
|
zgc:195050
|
zgc:195050 |
| chr16_-_42004544 | 0.55 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr17_-_6730247 | 0.54 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr24_-_40700596 | 0.54 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
| chr23_-_4925641 | 0.54 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr16_-_4640539 | 0.54 |
ENSDART00000076955
ENSDART00000131949 |
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr10_+_21563986 | 0.53 |
ENSDART00000100600
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr3_+_23703704 | 0.53 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr19_-_44054930 | 0.52 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr6_-_11768198 | 0.52 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr10_+_9281991 | 0.51 |
ENSDART00000139156
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr9_+_907459 | 0.51 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr24_-_33756003 | 0.51 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr25_+_13191615 | 0.51 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr23_+_45282858 | 0.50 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr10_+_21793670 | 0.50 |
ENSDART00000168918
|
pcdh1gc6
|
protocadherin 1 gamma c 6 |
| chr24_-_17049270 | 0.49 |
ENSDART00000175508
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr16_-_13789908 | 0.49 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr9_-_9989660 | 0.49 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr7_-_7845540 | 0.49 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr8_+_52619365 | 0.49 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr5_+_26795773 | 0.49 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr7_-_4296771 | 0.48 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr20_-_26039841 | 0.48 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr5_+_17727310 | 0.47 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr9_+_41080029 | 0.47 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr7_-_59564011 | 0.47 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr10_-_36808348 | 0.47 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr8_-_2434282 | 0.46 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr1_+_59090743 | 0.46 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr8_-_13972626 | 0.45 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_-_12930217 | 0.45 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr10_-_17103651 | 0.45 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr1_+_59067978 | 0.45 |
ENSDART00000172613
|
MFAP4 (1 of many)
|
si:ch1073-110a20.7 |
| chr19_+_5674907 | 0.45 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr12_+_6002715 | 0.44 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr5_-_55395384 | 0.44 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr13_+_23157053 | 0.44 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_+_44746154 | 0.43 |
ENSDART00000133926
|
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr17_-_31695217 | 0.43 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr15_-_29388012 | 0.43 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr8_+_29986265 | 0.43 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr21_-_22928214 | 0.43 |
ENSDART00000182760
|
dub
|
duboraya |
| chr20_+_6659770 | 0.42 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr9_-_34915351 | 0.42 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr12_+_31616412 | 0.42 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr17_-_16133249 | 0.42 |
ENSDART00000030919
|
pnoca
|
prepronociceptin a |
| chr25_-_18953322 | 0.42 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr13_-_9598320 | 0.41 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr13_+_39532050 | 0.41 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr17_+_30587333 | 0.41 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr25_+_1732838 | 0.41 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr4_-_12725513 | 0.41 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr5_+_39504136 | 0.41 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr12_-_26423439 | 0.40 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr1_-_59126139 | 0.40 |
ENSDART00000156105
|
si:ch1073-110a20.7
|
si:ch1073-110a20.7 |
| chr22_-_26175237 | 0.40 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr19_-_9712530 | 0.40 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr4_-_12723585 | 0.40 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr8_-_14151051 | 0.40 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr24_-_36727922 | 0.39 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr4_-_193762 | 0.39 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr21_+_22423286 | 0.39 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr5_+_4332220 | 0.39 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr11_-_6265574 | 0.39 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr15_+_28355023 | 0.39 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr12_+_17504559 | 0.38 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr3_-_57666996 | 0.38 |
ENSDART00000166855
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr22_-_3914162 | 0.38 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr23_-_4915118 | 0.38 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr12_+_22657925 | 0.38 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr6_+_8129543 | 0.38 |
ENSDART00000011724
|
klf1
|
Kruppel-like factor 1 (erythroid) |
| chr9_+_32301456 | 0.38 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr2_-_59145027 | 0.38 |
ENSDART00000128320
|
FO834803.1
|
|
| chr1_+_49568335 | 0.37 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr5_+_8919698 | 0.37 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr4_+_14957360 | 0.37 |
ENSDART00000002770
ENSDART00000111882 ENSDART00000148292 |
tspan33a
|
tetraspanin 33a |
| chr17_-_19534474 | 0.37 |
ENSDART00000192469
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr7_-_24491897 | 0.37 |
ENSDART00000173903
|
si:dkeyp-75h12.5
|
si:dkeyp-75h12.5 |
| chr22_+_3914318 | 0.37 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr9_+_38292947 | 0.37 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr12_-_31012741 | 0.36 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr21_-_22709251 | 0.36 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr16_-_8613499 | 0.36 |
ENSDART00000189189
|
cobl
|
cordon-bleu WH2 repeat protein |
| chr25_-_2081371 | 0.36 |
ENSDART00000104915
ENSDART00000156925 |
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr8_+_25351863 | 0.36 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr23_+_29889089 | 0.36 |
ENSDART00000149378
ENSDART00000089465 |
mxra8b
|
matrix-remodelling associated 8b |
| chr21_+_22846757 | 0.36 |
ENSDART00000185766
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr23_+_9220436 | 0.36 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr8_-_23684659 | 0.35 |
ENSDART00000136865
|
cfp
|
complement factor properdin |
| chr1_-_59139848 | 0.35 |
ENSDART00000191863
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr3_-_22212764 | 0.35 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr4_-_12914163 | 0.35 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr9_+_219124 | 0.34 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr19_-_10810006 | 0.34 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr14_-_2270973 | 0.34 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr5_-_20205075 | 0.34 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr25_-_31629095 | 0.34 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr22_+_14102397 | 0.34 |
ENSDART00000146397
ENSDART00000137530 |
he1.3
|
hatching enzyme 1, tandem duplicate 3 |
| chr9_-_5045378 | 0.33 |
ENSDART00000149268
|
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr19_+_43439457 | 0.33 |
ENSDART00000151571
|
ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr16_+_22587661 | 0.33 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr14_-_9281232 | 0.33 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr10_-_24343507 | 0.33 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr25_+_32755485 | 0.33 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr6_-_43449013 | 0.33 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr1_-_59134522 | 0.33 |
ENSDART00000152764
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr10_+_25982212 | 0.32 |
ENSDART00000128292
ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr16_+_50289916 | 0.32 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr3_-_18410968 | 0.32 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr11_+_37265692 | 0.32 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr23_-_9925568 | 0.32 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr14_+_21106444 | 0.32 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr22_-_18491813 | 0.32 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr14_+_21107032 | 0.32 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr25_+_32473277 | 0.31 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr8_+_471342 | 0.31 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr10_+_6496185 | 0.31 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr8_+_48480529 | 0.31 |
ENSDART00000186631
|
PRDM16
|
si:ch211-263k4.2 |
| chr22_+_30009926 | 0.31 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr4_+_12617108 | 0.31 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr8_-_14049404 | 0.31 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr16_+_12836143 | 0.31 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr20_+_2039518 | 0.31 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr2_+_6127593 | 0.31 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr5_-_39474235 | 0.31 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr12_+_25775734 | 0.31 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.6 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 1.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 2.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.7 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.4 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 1.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.3 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 1.9 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.7 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.4 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 0.3 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.9 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.9 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0060688 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0097240 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 1.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.7 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.2 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.5 | GO:0031638 | zymogen activation(GO:0031638) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 19.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 0.6 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.8 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 2.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 1.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |