Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
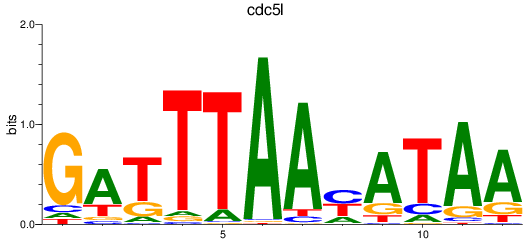
Results for cdc5l
Z-value: 2.28
Transcription factors associated with cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdc5l
|
ENSDARG00000043797 | CDC5 cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdc5l | dr11_v1_chr17_+_5061135_5061135 | 0.95 | 8.7e-05 | Click! |
Activity profile of cdc5l motif
Sorted Z-values of cdc5l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_35438300 | 3.53 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr6_+_40992409 | 2.93 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr9_+_44994214 | 2.91 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr22_+_25242322 | 2.63 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr22_+_25248961 | 2.61 |
ENSDART00000143079
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr6_+_40992883 | 2.45 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr23_-_44574059 | 2.43 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr22_+_25236657 | 2.35 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr9_+_34397516 | 2.31 |
ENSDART00000011304
ENSDART00000192973 |
med14
|
mediator complex subunit 14 |
| chr1_+_12195700 | 2.18 |
ENSDART00000040307
|
tdrd7a
|
tudor domain containing 7 a |
| chr16_+_29509133 | 2.16 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr2_-_57473980 | 2.16 |
ENSDART00000149353
ENSDART00000150034 |
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr10_+_37400838 | 2.07 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr19_+_27859546 | 2.05 |
ENSDART00000161908
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr19_-_27588842 | 1.97 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr24_-_9991153 | 1.92 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr5_+_66132394 | 1.90 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr3_-_15119856 | 1.85 |
ENSDART00000138328
|
xpo6
|
exportin 6 |
| chr11_+_6881001 | 1.83 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr8_-_16725573 | 1.80 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr25_+_17860798 | 1.79 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr9_+_34397843 | 1.77 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr19_-_27578929 | 1.73 |
ENSDART00000177368
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr10_-_21362320 | 1.70 |
ENSDART00000189789
|
avd
|
avidin |
| chr4_-_16124417 | 1.68 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr6_-_8704702 | 1.66 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr19_+_2835240 | 1.66 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr6_-_8466717 | 1.60 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr18_-_18874921 | 1.58 |
ENSDART00000193332
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr19_+_7001170 | 1.57 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr15_-_41689981 | 1.56 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr22_+_17261801 | 1.55 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr7_-_28647959 | 1.50 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr15_-_14038227 | 1.50 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr18_+_14633974 | 1.49 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr19_-_3056235 | 1.48 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr9_-_27398369 | 1.47 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr5_-_9216758 | 1.46 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr3_-_55537096 | 1.41 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr2_-_19234329 | 1.41 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr3_+_35542067 | 1.40 |
ENSDART00000146529
ENSDART00000084549 |
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr5_+_50879545 | 1.39 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr23_-_44577885 | 1.39 |
ENSDART00000166654
|
si:ch73-160p18.4
|
si:ch73-160p18.4 |
| chr3_+_29179329 | 1.36 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr17_+_43867889 | 1.35 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr24_-_10394277 | 1.35 |
ENSDART00000127568
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr3_-_54607166 | 1.34 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr24_-_38131381 | 1.33 |
ENSDART00000105666
|
crp6
|
C-reactive protein 6 |
| chr17_-_24937879 | 1.32 |
ENSDART00000153964
|
CR391986.1
|
|
| chr19_+_30450125 | 1.32 |
ENSDART00000073704
|
si:ch211-215a10.4
|
si:ch211-215a10.4 |
| chr2_-_23390779 | 1.30 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr15_+_20529197 | 1.28 |
ENSDART00000060935
ENSDART00000137926 ENSDART00000140087 |
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr20_-_27190393 | 1.27 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr14_-_30905963 | 1.27 |
ENSDART00000183543
ENSDART00000186441 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr11_-_33868881 | 1.26 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr19_+_7636941 | 1.26 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr16_-_21540077 | 1.26 |
ENSDART00000078790
|
stk31
|
serine/threonine kinase 31 |
| chr24_+_39129316 | 1.25 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr5_-_19006290 | 1.25 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr23_-_23179417 | 1.23 |
ENSDART00000122945
ENSDART00000091662 |
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr25_+_17860962 | 1.23 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr5_+_48666485 | 1.21 |
ENSDART00000158000
ENSDART00000031141 |
polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr20_+_42537768 | 1.21 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr17_-_19463355 | 1.21 |
ENSDART00000045881
|
dicer1
|
dicer 1, ribonuclease type III |
| chr15_+_12429206 | 1.20 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr20_+_43691208 | 1.20 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr8_+_10862353 | 1.19 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr2_-_22530969 | 1.18 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr1_-_53880639 | 1.18 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr10_-_32558917 | 1.18 |
ENSDART00000128888
ENSDART00000143301 |
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_+_24881680 | 1.17 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr12_-_18961289 | 1.17 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr13_-_25719628 | 1.17 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr6_+_40922572 | 1.17 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr4_+_9467049 | 1.17 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr18_+_18879733 | 1.16 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr25_+_19557628 | 1.16 |
ENSDART00000133859
|
lyrm5b
|
LYR motif containing 5b |
| chr25_-_3217115 | 1.15 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr20_-_38801981 | 1.15 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_6880725 | 1.15 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr23_-_24542156 | 1.14 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr19_+_24891747 | 1.14 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr25_+_17925424 | 1.13 |
ENSDART00000067305
|
zgc:103499
|
zgc:103499 |
| chr18_-_48296793 | 1.13 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr4_+_26056548 | 1.12 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr11_-_18283886 | 1.12 |
ENSDART00000019248
|
stimate
|
STIM activating enhance |
| chr7_+_13582256 | 1.11 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr8_-_35960987 | 1.11 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr6_-_10708960 | 1.08 |
ENSDART00000157704
|
si:dkey-34m19.3
|
si:dkey-34m19.3 |
| chr2_-_40890264 | 1.08 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr9_-_41040098 | 1.07 |
ENSDART00000008275
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr8_-_53044300 | 1.07 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr10_+_15454745 | 1.06 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr5_-_20135679 | 1.06 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr16_-_25741225 | 1.06 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr12_+_30367371 | 1.05 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr12_-_17863467 | 1.05 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr10_-_33251876 | 1.04 |
ENSDART00000184565
|
bcl7ba
|
BCL tumor suppressor 7Ba |
| chr6_+_60125033 | 1.04 |
ENSDART00000148557
ENSDART00000008224 |
aurka
|
aurora kinase A |
| chr24_+_21174851 | 1.04 |
ENSDART00000154940
ENSDART00000155977 ENSDART00000122762 |
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr23_-_18707418 | 1.03 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr21_+_20901505 | 1.02 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr4_-_77260727 | 1.01 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr6_-_12912606 | 1.01 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr12_+_1609563 | 1.01 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr2_-_7185460 | 1.01 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr16_-_42965192 | 1.01 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr23_+_24598910 | 1.00 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr23_+_26026383 | 1.00 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr21_-_25612658 | 1.00 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr4_-_13614797 | 1.00 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr12_-_611367 | 1.00 |
ENSDART00000152286
|
wu:fj29h11
|
wu:fj29h11 |
| chr7_+_13824150 | 1.00 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr19_+_40069524 | 1.00 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr21_-_19919020 | 0.99 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr24_+_24726956 | 0.99 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr21_+_27278120 | 0.98 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr9_+_38645136 | 0.98 |
ENSDART00000135505
|
slc12a8
|
solute carrier family 12, member 8 |
| chr6_-_32411703 | 0.97 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr9_+_38644976 | 0.96 |
ENSDART00000133849
ENSDART00000135774 |
slc12a8
|
solute carrier family 12, member 8 |
| chr23_-_37291793 | 0.96 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr20_+_22799857 | 0.96 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr8_+_52377516 | 0.95 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr22_-_9834944 | 0.95 |
ENSDART00000105944
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr12_+_20412564 | 0.94 |
ENSDART00000186783
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr1_-_30762264 | 0.94 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr3_-_61375496 | 0.94 |
ENSDART00000165188
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr3_-_31086770 | 0.93 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr21_-_18275226 | 0.93 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr17_-_10813885 | 0.92 |
ENSDART00000153913
|
fbxo34
|
F-box protein 34 |
| chr21_-_43457554 | 0.92 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr2_+_37295088 | 0.92 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr23_-_29553430 | 0.91 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr7_-_24994722 | 0.91 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr8_-_20245892 | 0.90 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr16_-_31622777 | 0.90 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr18_-_39200557 | 0.90 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr12_-_24812403 | 0.90 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr2_+_31806602 | 0.89 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr2_-_39558643 | 0.89 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr10_-_11840353 | 0.89 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr6_-_15492030 | 0.89 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr2_-_31700396 | 0.88 |
ENSDART00000142366
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr11_-_39118882 | 0.88 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr14_-_36763302 | 0.88 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr4_+_17642731 | 0.88 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr5_-_37117778 | 0.87 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_10634438 | 0.86 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr13_-_37474989 | 0.86 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr5_+_31779911 | 0.86 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr21_-_22357545 | 0.86 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_-_48188928 | 0.86 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr23_-_16981899 | 0.86 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr3_-_367283 | 0.85 |
ENSDART00000155936
ENSDART00000161964 ENSDART00000158560 ENSDART00000135595 ENSDART00000145890 |
mhc1zaa
|
major histocompatibility complex class I ZAA |
| chr20_-_15161669 | 0.85 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr2_+_52049239 | 0.84 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr10_-_28028998 | 0.84 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr7_-_14446512 | 0.82 |
ENSDART00000041577
|
kif7
|
kinesin family member 7 |
| chr8_+_13368150 | 0.82 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr10_-_26225548 | 0.82 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr8_-_14554785 | 0.82 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_-_25366541 | 0.81 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr23_+_4348479 | 0.81 |
ENSDART00000182425
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr10_+_3428194 | 0.81 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr17_-_14774117 | 0.81 |
ENSDART00000080401
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr23_+_24589633 | 0.80 |
ENSDART00000143694
|
mlh3
|
mutL homolog 3 (E. coli) |
| chr17_-_8312923 | 0.80 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr3_-_34586403 | 0.80 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr19_+_15444210 | 0.80 |
ENSDART00000142509
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr20_+_28803642 | 0.80 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr16_-_12060770 | 0.79 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr8_-_29930821 | 0.79 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr3_-_18189283 | 0.79 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr17_+_16090436 | 0.79 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr5_-_15494164 | 0.79 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr11_-_16395956 | 0.78 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr22_+_23430688 | 0.78 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr6_-_37745508 | 0.78 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr20_-_34069956 | 0.78 |
ENSDART00000017941
|
tprb
|
translocated promoter region b, nuclear basket protein |
| chr15_-_41689684 | 0.78 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr23_+_4348265 | 0.78 |
ENSDART00000136287
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr17_-_26537928 | 0.78 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr3_-_61385117 | 0.77 |
ENSDART00000170164
|
znf1028
|
zinc finger protein 1028 |
| chr5_-_40024902 | 0.77 |
ENSDART00000017451
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr19_+_3056450 | 0.77 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_7897563 | 0.77 |
ENSDART00000185232
|
ubn2b
|
ubinuclein 2b |
| chr18_-_18875308 | 0.76 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr25_-_12805295 | 0.76 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr25_-_6432463 | 0.76 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr17_-_6954719 | 0.76 |
ENSDART00000188180
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr10_+_6907715 | 0.76 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr20_+_26394324 | 0.76 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr3_+_26244353 | 0.76 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr6_+_26948093 | 0.75 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr24_-_31904924 | 0.75 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr7_+_55314206 | 0.75 |
ENSDART00000132852
|
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr16_-_26528140 | 0.75 |
ENSDART00000134448
ENSDART00000147062 |
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr20_-_23876291 | 0.75 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr18_-_17087138 | 0.75 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr16_+_29690708 | 0.75 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdc5l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.6 | 6.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.5 | 1.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 2.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.4 | 1.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.4 | 1.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.3 | 1.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 1.0 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 1.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 0.5 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.0 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 0.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.7 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 0.9 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 2.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 0.6 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.2 | 2.5 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.2 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.5 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.2 | 0.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.5 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.9 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 2.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 1.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.4 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 1.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 0.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 4.1 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 1.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.2 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 1.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.5 | GO:0019068 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 1.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 2.0 | GO:0034661 | ncRNA catabolic process(GO:0034661) |
| 0.1 | 0.6 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 0.7 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.4 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.5 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 1.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.9 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.7 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.1 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.8 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 1.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.3 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.7 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.5 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 1.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.1 | GO:0051785 | positive regulation of mitotic nuclear division(GO:0045840) positive regulation of nuclear division(GO:0051785) |
| 0.1 | 0.4 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.5 | GO:0010522 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.1 | 0.2 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.6 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 3.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 2.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.8 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.9 | GO:0043506 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.3 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 1.6 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 2.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.5 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.1 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 2.0 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.9 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.1 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 1.9 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.3 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.5 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.6 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.8 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 1.2 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.2 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 5.6 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.6 | 4.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.5 | 1.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 1.4 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.4 | 1.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 1.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 1.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 1.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 0.8 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 0.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.8 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 0.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 0.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 1.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.2 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 4.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 3.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 4.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 4.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.3 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 2.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 1.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 1.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.4 | 1.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.3 | 1.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.3 | 1.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.3 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.3 | 1.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.3 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 5.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.9 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.6 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.9 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 0.7 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 2.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 0.5 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.5 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 0.6 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.4 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.6 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 2.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.3 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 4.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.4 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.2 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 2.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 2.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 8.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 5.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.0 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 6.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |