Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
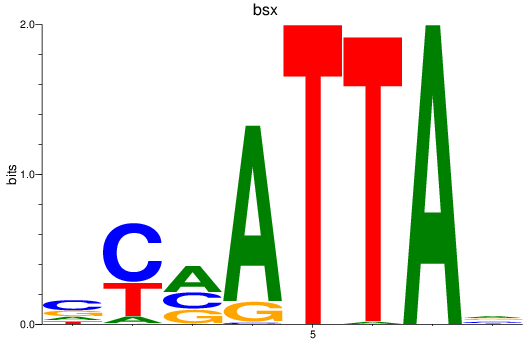
Results for bsx
Z-value: 0.52
Transcription factors associated with bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bsx
|
ENSDARG00000068976 | brain-specific homeobox |
|
bsx
|
ENSDARG00000110782 | brain-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bsx | dr11_v1_chr10_-_29892486_29892486 | -0.54 | 1.4e-01 | Click! |
Activity profile of bsx motif
Sorted Z-values of bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_30452945 | 0.65 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr17_-_48944465 | 0.57 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr11_-_1550709 | 0.52 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr8_-_20243389 | 0.52 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr18_-_33979693 | 0.48 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr11_-_6452444 | 0.43 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr2_+_41926707 | 0.43 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr14_+_34490445 | 0.42 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_-_23426339 | 0.41 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr21_+_25777425 | 0.41 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_44560165 | 0.40 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr12_-_42368296 | 0.38 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr24_-_10014512 | 0.37 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr23_-_25098913 | 0.37 |
ENSDART00000137316
|
CR847803.1
|
|
| chr24_+_12835935 | 0.36 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr10_-_21362320 | 0.36 |
ENSDART00000189789
|
avd
|
avidin |
| chr11_+_18130300 | 0.35 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr5_+_68807170 | 0.35 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr10_-_21362071 | 0.34 |
ENSDART00000125167
|
avd
|
avidin |
| chr2_-_38287987 | 0.34 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr4_-_49069087 | 0.34 |
ENSDART00000150622
|
si:ch211-234c11.3
|
si:ch211-234c11.3 |
| chr19_-_20403507 | 0.34 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr16_+_29509133 | 0.33 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_-_13149453 | 0.33 |
ENSDART00000172578
|
si:dkey-228b2.6
|
si:dkey-228b2.6 |
| chr10_+_6884627 | 0.31 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr8_-_554540 | 0.31 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr10_-_25217347 | 0.31 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr19_-_20403845 | 0.31 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr2_+_33326522 | 0.30 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr21_-_43666420 | 0.30 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr1_-_55248496 | 0.30 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr4_-_55039830 | 0.29 |
ENSDART00000170096
|
si:dkey-56m15.3
|
si:dkey-56m15.3 |
| chr11_+_18175893 | 0.29 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr3_+_18807006 | 0.28 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr19_+_15440841 | 0.28 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr13_-_21660203 | 0.27 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr24_-_10021341 | 0.26 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr11_+_18157260 | 0.26 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr5_+_50879545 | 0.25 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr19_+_42470396 | 0.25 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr10_+_6884123 | 0.24 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr3_-_3448095 | 0.24 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr3_-_53091946 | 0.24 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr25_-_29074064 | 0.23 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_15441022 | 0.23 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr24_-_4450238 | 0.23 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr5_-_9216758 | 0.23 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr21_-_37973819 | 0.23 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr10_-_34002185 | 0.23 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_20793982 | 0.21 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr11_+_18183220 | 0.21 |
ENSDART00000113468
|
LO018315.10
|
|
| chr23_+_38251864 | 0.21 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr17_-_21162821 | 0.21 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr21_-_22547496 | 0.21 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr20_-_48898560 | 0.20 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr19_+_31585917 | 0.19 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr11_-_45138857 | 0.19 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr14_-_28567845 | 0.19 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr1_-_33645967 | 0.19 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr21_-_37194365 | 0.19 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr22_+_18315490 | 0.19 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr3_+_13848226 | 0.19 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr5_-_68333081 | 0.19 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr6_+_40922572 | 0.19 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_+_41191482 | 0.19 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr7_+_24528430 | 0.18 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr17_+_16046314 | 0.18 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_-_40194732 | 0.18 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr20_+_46513651 | 0.18 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr5_-_19006290 | 0.18 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr6_+_28208973 | 0.17 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr1_+_23563691 | 0.17 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr10_-_33297864 | 0.17 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr5_-_12063381 | 0.17 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr3_-_53092509 | 0.17 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr20_+_18260358 | 0.17 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_63375620 | 0.17 |
ENSDART00000185568
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr23_-_31913069 | 0.17 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr4_+_9177997 | 0.16 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr21_-_2322102 | 0.16 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr16_+_28994709 | 0.16 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr6_-_43283122 | 0.16 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr23_-_25135046 | 0.16 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr20_-_48898371 | 0.16 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr2_+_41526904 | 0.16 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr3_+_26244353 | 0.16 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr7_-_54679595 | 0.16 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr17_+_16046132 | 0.16 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr14_-_24110251 | 0.16 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr10_-_35257458 | 0.16 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr8_+_41037541 | 0.16 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr6_+_50393047 | 0.15 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr17_+_24821627 | 0.15 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr24_+_1023839 | 0.15 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr5_+_63375386 | 0.15 |
ENSDART00000137855
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr24_+_26134209 | 0.15 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr24_-_30843250 | 0.15 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr4_+_52356485 | 0.15 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr9_-_27748868 | 0.14 |
ENSDART00000190306
|
tbccd1
|
TBCC domain containing 1 |
| chr3_+_1637358 | 0.14 |
ENSDART00000180266
|
CR394546.5
|
|
| chr2_-_42071558 | 0.14 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr23_-_29357764 | 0.14 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr24_+_24170914 | 0.14 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr2_+_56891858 | 0.14 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr13_+_36984794 | 0.14 |
ENSDART00000137328
|
frmd6
|
FERM domain containing 6 |
| chr2_+_8780443 | 0.14 |
ENSDART00000137768
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr16_+_16266428 | 0.14 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr9_+_11034314 | 0.14 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr6_-_35046735 | 0.14 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr21_-_3672343 | 0.14 |
ENSDART00000086492
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr10_-_13343831 | 0.14 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr11_-_18557929 | 0.13 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr5_+_6954162 | 0.13 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr19_-_20403318 | 0.13 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr12_+_10631266 | 0.13 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr25_-_36370292 | 0.13 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr2_-_37103622 | 0.13 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr3_-_26244256 | 0.13 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr17_+_28102487 | 0.13 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr4_-_14191434 | 0.13 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr5_-_68779747 | 0.13 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr6_-_13022166 | 0.13 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr14_+_24845941 | 0.13 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr23_+_38957472 | 0.13 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr6_-_41138854 | 0.13 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr10_+_17371356 | 0.12 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr18_+_17537344 | 0.12 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr2_-_32387441 | 0.12 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr18_-_8857137 | 0.12 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr4_+_13586689 | 0.12 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr20_-_28433990 | 0.12 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr23_+_33947874 | 0.12 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr14_+_38849432 | 0.12 |
ENSDART00000052511
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr24_+_37800102 | 0.12 |
ENSDART00000187591
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr13_-_11984867 | 0.12 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr2_-_26596794 | 0.12 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr9_-_21460164 | 0.12 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr24_+_34069675 | 0.12 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr2_-_57076687 | 0.12 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr8_-_1838315 | 0.12 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr24_-_2450597 | 0.12 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr21_+_33187992 | 0.11 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr9_-_40873934 | 0.11 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr4_-_43388943 | 0.11 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr12_+_30367371 | 0.11 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr19_+_16015881 | 0.11 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr20_-_7133546 | 0.11 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr11_+_2458264 | 0.11 |
ENSDART00000188341
|
tp53rk
|
TP53 regulating kinase |
| chr15_-_14884332 | 0.11 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr9_+_34331368 | 0.11 |
ENSDART00000147913
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr11_+_11303458 | 0.11 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr12_+_16087077 | 0.11 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr17_-_49412313 | 0.11 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr16_-_41714988 | 0.11 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr12_+_32073660 | 0.11 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr3_-_43650189 | 0.11 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr7_+_17908235 | 0.11 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr2_-_32666174 | 0.11 |
ENSDART00000133660
|
puf60a
|
poly-U binding splicing factor a |
| chr3_+_39656188 | 0.11 |
ENSDART00000186292
ENSDART00000156038 |
epn2
|
epsin 2 |
| chr13_+_17468161 | 0.11 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr22_-_18387059 | 0.11 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr7_-_51368681 | 0.11 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr6_-_39649504 | 0.10 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr6_-_40922971 | 0.10 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr9_+_29548195 | 0.10 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr18_+_50526290 | 0.10 |
ENSDART00000175194
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr5_-_41142467 | 0.10 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr23_+_38957738 | 0.10 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr24_-_25166720 | 0.10 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr7_+_34592526 | 0.10 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_+_24287927 | 0.10 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr21_-_11654422 | 0.10 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr23_-_31913231 | 0.10 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr19_+_3056450 | 0.10 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr15_+_41027466 | 0.10 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr23_-_18913032 | 0.10 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr10_-_23099809 | 0.10 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr11_+_39928828 | 0.10 |
ENSDART00000137516
ENSDART00000134082 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr22_+_2260497 | 0.10 |
ENSDART00000142890
|
si:dkey-4c15.9
|
si:dkey-4c15.9 |
| chr1_-_513762 | 0.10 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr12_+_30367079 | 0.10 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr16_-_17347727 | 0.10 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr11_+_44226200 | 0.10 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr25_+_29662411 | 0.10 |
ENSDART00000077445
|
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr8_+_8166285 | 0.10 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr13_-_12005429 | 0.10 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr21_+_38732945 | 0.09 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr19_+_7152966 | 0.09 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr11_+_30000814 | 0.09 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr19_-_6774871 | 0.09 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr7_+_29863548 | 0.09 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr18_+_17538165 | 0.09 |
ENSDART00000147839
|
nup93
|
nucleoporin 93 |
| chr6_-_25163722 | 0.09 |
ENSDART00000192225
|
znf326
|
zinc finger protein 326 |
| chr15_+_29662401 | 0.09 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr6_+_13024448 | 0.09 |
ENSDART00000104768
|
pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr22_+_9003090 | 0.09 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr6_+_39098397 | 0.09 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr17_+_10501647 | 0.09 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr10_+_33744098 | 0.09 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr2_+_42072231 | 0.09 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.8 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.2 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.5 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.6 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.6 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.3 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |