Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
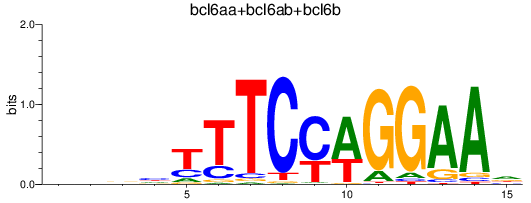
Results for bcl6aa+bcl6ab+bcl6b
Z-value: 0.72
Transcription factors associated with bcl6aa+bcl6ab+bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl6ab
|
ENSDARG00000069295 | BCL6A transcription repressor b |
|
bcl6b
|
ENSDARG00000069335 | BCL6B transcription repressor |
|
bcl6aa
|
ENSDARG00000070864 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000111395 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000112502 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000116260 | BCL6A transcription repressor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl6a | dr11_v1_chr6_-_28222592_28222592 | -0.66 | 5.4e-02 | Click! |
| bcl6b | dr11_v1_chr7_+_20017211_20017211 | -0.59 | 9.1e-02 | Click! |
| bcl6ab | dr11_v1_chr2_-_10098191_10098191 | -0.13 | 7.5e-01 | Click! |
Activity profile of bcl6aa+bcl6ab+bcl6b motif
Sorted Z-values of bcl6aa+bcl6ab+bcl6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_10166297 | 0.76 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr9_-_43644261 | 0.59 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr25_-_29415369 | 0.53 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr8_-_23240156 | 0.49 |
ENSDART00000131632
|
ptk6a
|
PTK6 protein tyrosine kinase 6a |
| chr4_+_19127973 | 0.47 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr18_-_46183462 | 0.46 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr11_-_8167799 | 0.45 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr2_-_38284648 | 0.45 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr3_+_40289418 | 0.44 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr23_-_14830627 | 0.41 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr13_-_37608441 | 0.40 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr18_+_22109379 | 0.36 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr23_+_1033668 | 0.35 |
ENSDART00000143391
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr7_-_58776400 | 0.35 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr9_+_20869166 | 0.35 |
ENSDART00000147892
|
wdr3
|
WD repeat domain 3 |
| chr9_-_9225980 | 0.34 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr22_+_7480465 | 0.34 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr7_+_5529967 | 0.34 |
ENSDART00000142709
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr10_-_3332362 | 0.34 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr8_+_1189798 | 0.33 |
ENSDART00000193474
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr2_+_13710439 | 0.33 |
ENSDART00000155712
|
ebna1bp2
|
EBNA1 binding protein 2 |
| chr8_-_7232413 | 0.33 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr21_+_15870752 | 0.33 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr14_-_28567845 | 0.31 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr22_+_25289360 | 0.31 |
ENSDART00000143782
|
si:ch211-154a22.8
|
si:ch211-154a22.8 |
| chr21_+_26720803 | 0.31 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr8_-_23599096 | 0.30 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr11_-_22303678 | 0.30 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr23_-_33558161 | 0.30 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr14_-_6987649 | 0.30 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr9_+_40825065 | 0.29 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr13_-_31349284 | 0.29 |
ENSDART00000145887
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr11_-_11791718 | 0.29 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr10_-_29827000 | 0.29 |
ENSDART00000131418
|
zpr1
|
ZPR1 zinc finger |
| chr24_+_31361407 | 0.29 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr2_-_9989919 | 0.27 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr3_-_6608342 | 0.27 |
ENSDART00000161188
|
si:ch73-59f11.3
|
si:ch73-59f11.3 |
| chr23_+_42482137 | 0.27 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr2_-_38125657 | 0.26 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr11_+_43375326 | 0.26 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr17_+_16046314 | 0.26 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_+_26804197 | 0.26 |
ENSDART00000135688
|
si:dkey-44g23.2
|
si:dkey-44g23.2 |
| chr13_+_33688474 | 0.26 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr18_+_45228691 | 0.25 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr9_+_29430432 | 0.25 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr13_-_37631092 | 0.25 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr2_+_327081 | 0.25 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr16_+_40954481 | 0.25 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr24_-_2829049 | 0.24 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr7_+_38809241 | 0.24 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr17_-_21278582 | 0.24 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr17_+_24597001 | 0.24 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr1_+_35495368 | 0.24 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr5_+_36895545 | 0.24 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr17_-_21278846 | 0.24 |
ENSDART00000181356
|
hspa12a
|
heat shock protein 12A |
| chr5_-_25582721 | 0.23 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr9_-_12401871 | 0.23 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr14_-_38828057 | 0.23 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr6_+_37655078 | 0.23 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr18_-_8378251 | 0.23 |
ENSDART00000192807
|
sephs1
|
selenophosphate synthetase 1 |
| chr15_-_40246396 | 0.23 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr2_-_57344037 | 0.23 |
ENSDART00000148873
|
tcf3a
|
transcription factor 3a |
| chr20_-_34767472 | 0.23 |
ENSDART00000186710
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr1_-_9195629 | 0.22 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr23_+_42396934 | 0.22 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr21_-_30026359 | 0.22 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr23_+_4350897 | 0.22 |
ENSDART00000190593
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr8_+_22359881 | 0.22 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr15_+_880751 | 0.22 |
ENSDART00000156724
|
si:dkey-7i4.10
|
si:dkey-7i4.10 |
| chr5_+_29160324 | 0.22 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr6_+_26948093 | 0.22 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr14_-_21123325 | 0.22 |
ENSDART00000180889
ENSDART00000188539 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr19_-_8798178 | 0.21 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr3_+_39656188 | 0.21 |
ENSDART00000186292
ENSDART00000156038 |
epn2
|
epsin 2 |
| chr20_-_37813863 | 0.21 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr3_-_60602432 | 0.20 |
ENSDART00000163235
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr5_+_50953240 | 0.20 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr20_+_25625872 | 0.20 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr20_-_33704753 | 0.20 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_-_29195063 | 0.20 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr11_+_19603251 | 0.20 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr7_-_26627252 | 0.19 |
ENSDART00000164050
ENSDART00000159826 |
phf23b
|
PHD finger protein 23b |
| chr18_+_14684115 | 0.19 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr22_+_26400519 | 0.19 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr17_+_16046132 | 0.19 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_-_2297832 | 0.19 |
ENSDART00000158266
|
znf740a
|
zinc finger protein 740a |
| chr12_-_1031970 | 0.19 |
ENSDART00000105292
|
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr15_+_20239141 | 0.18 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr10_-_42147318 | 0.18 |
ENSDART00000134890
|
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr1_-_50791280 | 0.18 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr16_+_16849220 | 0.18 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr23_-_19682971 | 0.18 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr1_-_20068155 | 0.18 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr12_-_33972798 | 0.18 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr22_+_3238474 | 0.17 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr17_-_31308658 | 0.17 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr17_+_4368859 | 0.17 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr5_+_36895860 | 0.17 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr6_-_12296170 | 0.16 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr24_+_39027481 | 0.16 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr2_+_26237322 | 0.16 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr12_-_4243268 | 0.16 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr23_+_42346799 | 0.16 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr17_+_34206167 | 0.16 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr19_-_47782916 | 0.16 |
ENSDART00000063337
|
cdca8
|
cell division cycle associated 8 |
| chr20_+_39457598 | 0.16 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr25_-_13408760 | 0.16 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr16_-_45917322 | 0.16 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr10_+_40324395 | 0.16 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr16_-_41667101 | 0.16 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr19_-_30420602 | 0.15 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr3_-_3703572 | 0.15 |
ENSDART00000111017
|
si:ch211-163m16.7
|
si:ch211-163m16.7 |
| chr19_-_20777351 | 0.15 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr6_+_13046720 | 0.15 |
ENSDART00000165896
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_+_23808640 | 0.15 |
ENSDART00000024619
|
gorasp1a
|
golgi reassembly stacking protein 1a |
| chr16_+_10330178 | 0.15 |
ENSDART00000121966
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr11_+_10548171 | 0.15 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr2_+_44972720 | 0.15 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr16_+_10329701 | 0.15 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr21_-_2238277 | 0.15 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr9_-_12659140 | 0.15 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr17_-_30521043 | 0.15 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr16_+_42772678 | 0.15 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr19_+_43885770 | 0.15 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr21_+_34122801 | 0.14 |
ENSDART00000182627
|
hmgb3b
|
high mobility group box 3b |
| chr17_-_45378473 | 0.14 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr7_+_26029672 | 0.14 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr10_-_39281475 | 0.14 |
ENSDART00000175136
|
cry5
|
cryptochrome circadian clock 5 |
| chr25_+_16915974 | 0.14 |
ENSDART00000188923
|
zgc:77158
|
zgc:77158 |
| chr22_+_1462177 | 0.14 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr18_-_20444296 | 0.14 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr2_-_7185460 | 0.14 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr1_-_56596701 | 0.14 |
ENSDART00000133693
|
LO018605.1
|
|
| chr13_+_25549425 | 0.14 |
ENSDART00000087553
ENSDART00000169199 |
sec23ip
|
SEC23 interacting protein |
| chr8_+_11687254 | 0.14 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr5_-_68495224 | 0.13 |
ENSDART00000187955
|
ephb4a
|
eph receptor B4a |
| chr5_-_25066780 | 0.13 |
ENSDART00000002118
ENSDART00000182575 |
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr23_+_45229198 | 0.13 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr9_-_55772937 | 0.13 |
ENSDART00000159192
|
akap17a
|
A kinase (PRKA) anchor protein 17A |
| chr21_+_17541634 | 0.13 |
ENSDART00000191700
ENSDART00000163238 |
stom
|
stomatin |
| chr3_+_24511959 | 0.13 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr5_+_28849155 | 0.13 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr8_-_23783633 | 0.13 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr10_+_585719 | 0.13 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr2_-_9744081 | 0.13 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr15_+_22333778 | 0.13 |
ENSDART00000114937
|
pde9al
|
phosphodiesterase 9A like |
| chr7_-_54320088 | 0.13 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr2_+_45100963 | 0.13 |
ENSDART00000160122
|
CU929150.2
|
|
| chr3_+_50172452 | 0.13 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr19_+_47783137 | 0.13 |
ENSDART00000024777
ENSDART00000158979 |
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr8_+_47099033 | 0.13 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr20_+_27087539 | 0.13 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr3_+_62236443 | 0.13 |
ENSDART00000180482
ENSDART00000193718 |
BX470259.4
|
|
| chr22_-_22242884 | 0.12 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr6_-_11800427 | 0.12 |
ENSDART00000126243
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr25_-_17395315 | 0.12 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr11_-_38914265 | 0.12 |
ENSDART00000141229
|
si:ch211-122l14.4
|
si:ch211-122l14.4 |
| chr10_-_40557210 | 0.12 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr7_+_26549846 | 0.12 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr19_+_14352332 | 0.12 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr9_-_27738110 | 0.12 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr25_+_11281970 | 0.12 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr7_-_22941472 | 0.12 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr1_-_44928987 | 0.12 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr17_-_28811747 | 0.12 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr1_+_10318089 | 0.12 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr6_-_7726849 | 0.12 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr12_-_13337033 | 0.12 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr18_+_3243292 | 0.12 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr14_+_7932973 | 0.11 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr4_+_76306625 | 0.11 |
ENSDART00000193823
ENSDART00000180622 |
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr13_-_8888334 | 0.11 |
ENSDART00000059881
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr3_-_7147226 | 0.11 |
ENSDART00000178155
|
BX005085.5
|
|
| chr11_-_43226255 | 0.11 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_+_32492467 | 0.11 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr14_+_29945070 | 0.11 |
ENSDART00000185039
|
fam149a
|
family with sequence similarity 149 member A |
| chr1_-_47114310 | 0.11 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr7_+_12950507 | 0.11 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr11_-_40742424 | 0.11 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr8_-_19246342 | 0.11 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr3_+_62219158 | 0.11 |
ENSDART00000162354
|
BX470259.5
|
|
| chr3_+_35608385 | 0.11 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr10_+_40284003 | 0.11 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr11_-_287670 | 0.11 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr3_+_12861158 | 0.11 |
ENSDART00000171237
|
kcnj2b
|
potassium inwardly-rectifying channel, subfamily J, member 2b |
| chr3_-_50998577 | 0.11 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr24_+_10397865 | 0.11 |
ENSDART00000155557
|
si:ch211-69l10.4
|
si:ch211-69l10.4 |
| chr17_+_7522777 | 0.11 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr3_-_52661242 | 0.11 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr19_+_28291376 | 0.11 |
ENSDART00000139433
ENSDART00000103855 |
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr3_-_60589292 | 0.10 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr16_+_25343934 | 0.10 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr8_+_23711842 | 0.10 |
ENSDART00000128783
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr4_-_73739119 | 0.10 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr3_+_60589157 | 0.10 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr3_-_30888415 | 0.10 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr7_+_14005111 | 0.10 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr23_+_42292748 | 0.10 |
ENSDART00000166113
ENSDART00000158684 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr23_-_1571682 | 0.10 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr20_+_25581627 | 0.10 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl6aa+bcl6ab+bcl6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 0.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.3 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.3 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.2 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.2 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.0 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:0033028 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.3 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0002370 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) nuclear periphery(GO:0034399) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 1.2 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |