Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
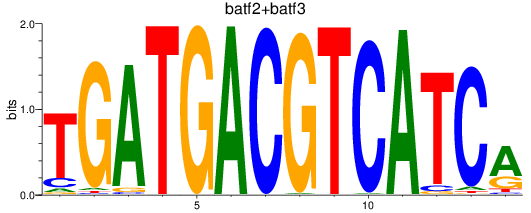
Results for batf2+batf3
Z-value: 1.37
Transcription factors associated with batf2+batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
batf3
|
ENSDARG00000042577 | basic leucine zipper transcription factor, ATF-like 3 |
|
batf2
|
ENSDARG00000105562 | si_dkey-23i12.7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf3 | dr11_v1_chr20_-_37813863_37813863 | 0.94 | 2.0e-04 | Click! |
| si:dkey-23i12.7 | dr11_v1_chr7_+_25049545_25049577 | -0.16 | 6.7e-01 | Click! |
Activity profile of batf2+batf3 motif
Sorted Z-values of batf2+batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_29387215 | 3.52 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr16_+_27614989 | 3.34 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr23_-_1571682 | 2.37 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr15_+_34963316 | 2.17 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr25_-_6447835 | 2.10 |
ENSDART00000012820
|
snupn
|
snurportin 1 |
| chr3_-_40658820 | 2.04 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr6_+_475264 | 1.89 |
ENSDART00000193615
|
LO017974.1
|
|
| chr17_+_15674052 | 1.89 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr16_-_41667101 | 1.84 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr25_-_6448050 | 1.82 |
ENSDART00000180616
|
snupn
|
snurportin 1 |
| chr7_-_69352424 | 1.62 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr25_+_19913611 | 1.62 |
ENSDART00000155029
|
gramd4b
|
GRAM domain containing 4b |
| chr8_+_11687586 | 1.60 |
ENSDART00000146241
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr11_+_24314148 | 1.33 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_28166165 | 1.25 |
ENSDART00000169360
ENSDART00000192311 |
ephb2b
|
eph receptor B2b |
| chr14_-_32959851 | 1.24 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr21_-_14762944 | 1.19 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr11_+_24313931 | 1.10 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr15_-_20412286 | 1.03 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr22_+_1821718 | 0.94 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr22_+_2228919 | 0.93 |
ENSDART00000133475
|
znf1161
|
zinc finger protein 1161 |
| chr22_+_1517318 | 0.90 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr22_+_2648931 | 0.89 |
ENSDART00000106357
|
zgc:110821
|
zgc:110821 |
| chr3_-_16413606 | 0.89 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr22_+_1641547 | 0.88 |
ENSDART00000159804
|
si:dkey-1b17.2
|
si:dkey-1b17.2 |
| chr22_+_1462177 | 0.85 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr22_+_2715815 | 0.83 |
ENSDART00000042770
|
znf1163
|
zinc finger protein 1163 |
| chr1_+_41666611 | 0.82 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr1_-_20068155 | 0.81 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr22_+_1751640 | 0.79 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr22_+_1668471 | 0.77 |
ENSDART00000163358
|
si:dkey-1b17.5
|
si:dkey-1b17.5 |
| chr22_+_1953096 | 0.77 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr22_+_1940595 | 0.77 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr10_+_28306749 | 0.77 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr5_+_11812089 | 0.76 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr22_+_2524615 | 0.76 |
ENSDART00000134277
|
znf1003
|
zinc finger protein 1003 |
| chr2_+_27341559 | 0.75 |
ENSDART00000176838
|
tesk2
|
testis-specific kinase 2 |
| chr24_+_25822999 | 0.75 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr22_+_2751887 | 0.74 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr1_+_10305611 | 0.67 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr22_+_2315996 | 0.66 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr17_-_14591764 | 0.66 |
ENSDART00000174703
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr22_+_2403068 | 0.65 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr4_-_13502549 | 0.65 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr25_+_37435720 | 0.61 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr11_-_43226255 | 0.60 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr22_+_2345752 | 0.54 |
ENSDART00000143687
|
zgc:174224
|
zgc:174224 |
| chr5_+_20823409 | 0.52 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr18_-_12451772 | 0.52 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr3_-_40162843 | 0.51 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr8_-_21110233 | 0.49 |
ENSDART00000127371
ENSDART00000100276 |
tmco1
|
transmembrane and coiled-coil domains 1 |
| chr22_+_1904546 | 0.49 |
ENSDART00000164865
|
si:dkey-15h8.10
|
si:dkey-15h8.10 |
| chr22_+_2390544 | 0.48 |
ENSDART00000147345
ENSDART00000133611 |
zgc:171435
|
zgc:171435 |
| chr22_+_2183110 | 0.47 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr22_+_2207502 | 0.46 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr22_+_2801464 | 0.46 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr22_+_2120815 | 0.38 |
ENSDART00000169133
ENSDART00000166644 ENSDART00000162221 ENSDART00000157840 |
znf1155
|
zinc finger protein 1155 |
| chr7_+_8670398 | 0.38 |
ENSDART00000164195
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr22_+_2298419 | 0.37 |
ENSDART00000143499
|
znf1180
|
zinc finger protein 1180 |
| chr22_+_1947494 | 0.37 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr22_+_2111120 | 0.33 |
ENSDART00000165525
|
znf1144
|
zinc finger protein 1144 |
| chr3_-_49514874 | 0.32 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr22_+_2819613 | 0.32 |
ENSDART00000131234
|
si:dkey-20i20.3
|
si:dkey-20i20.3 |
| chr22_+_2254972 | 0.28 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr22_+_2189356 | 0.28 |
ENSDART00000157502
ENSDART00000164188 |
znf1165
|
zinc finger protein 1165 |
| chr22_+_2844865 | 0.26 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr7_+_13464878 | 0.26 |
ENSDART00000172969
|
dagla
|
diacylglycerol lipase, alpha |
| chr22_+_1922293 | 0.18 |
ENSDART00000165758
|
znf1181
|
zinc finger protein 1181 |
| chr22_+_1615691 | 0.18 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr15_-_762319 | 0.17 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr22_+_1930589 | 0.15 |
ENSDART00000159807
|
znf1153
|
zinc finger protein 1153 |
| chr22_+_2247645 | 0.11 |
ENSDART00000143366
ENSDART00000147852 |
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr8_+_53120278 | 0.11 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr22_+_1853999 | 0.10 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr25_+_7321675 | 0.06 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr4_+_1283068 | 0.03 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of batf2+batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.5 | 2.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.6 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.4 | 1.8 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.8 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 1.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 5.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 1.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.7 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.8 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 0.9 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 0.8 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.2 | 1.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 3.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 3.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |