Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
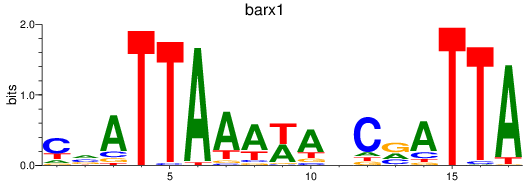
Results for barx1
Z-value: 1.00
Transcription factors associated with barx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx1
|
ENSDARG00000007407 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx1 | dr11_v1_chr11_+_27543093_27543093 | 0.57 | 1.1e-01 | Click! |
Activity profile of barx1 motif
Sorted Z-values of barx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 3.72 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr16_-_55028740 | 2.41 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr23_+_20422661 | 2.22 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr14_+_46313396 | 1.78 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_+_32206378 | 1.75 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr3_+_60721342 | 1.67 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr13_+_1726953 | 1.66 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr25_+_20089986 | 1.40 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr18_-_49318823 | 1.37 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr4_+_9669717 | 1.37 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr2_-_7666021 | 1.33 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr2_+_22795494 | 1.31 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr21_+_39185461 | 1.25 |
ENSDART00000178419
|
cryba1b
|
crystallin, beta A1b |
| chr14_-_26536504 | 1.19 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr3_-_55128258 | 1.18 |
ENSDART00000101734
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr5_-_66702479 | 1.16 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr2_-_31767827 | 1.13 |
ENSDART00000114928
|
and2
|
actinodin2 |
| chr12_-_16898140 | 1.10 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr5_+_24305877 | 1.08 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr5_-_41531629 | 1.07 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr12_-_16923162 | 1.06 |
ENSDART00000106072
|
si:dkey-26g8.5
|
si:dkey-26g8.5 |
| chr1_+_53374454 | 1.04 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr17_-_29224908 | 0.99 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr12_+_27141140 | 0.96 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr17_+_4030493 | 0.96 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr14_+_46313135 | 0.92 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_-_61205711 | 0.91 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr22_+_26400519 | 0.90 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr22_+_12770877 | 0.86 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr9_-_42484444 | 0.83 |
ENSDART00000048320
ENSDART00000047653 |
tfpia
|
tissue factor pathway inhibitor a |
| chr23_-_21453614 | 0.83 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr19_+_8973042 | 0.81 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr14_+_7140997 | 0.81 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr19_-_10554240 | 0.80 |
ENSDART00000104539
ENSDART00000144923 |
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr14_-_25599002 | 0.80 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr1_+_59293873 | 0.79 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr9_-_22135576 | 0.79 |
ENSDART00000101902
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr7_+_22675475 | 0.79 |
ENSDART00000134423
|
ponzr3
|
plac8 onzin related protein 3 |
| chr10_+_1396940 | 0.79 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr4_+_76575585 | 0.79 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr22_+_10651726 | 0.78 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr12_-_16720432 | 0.77 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr22_-_15587360 | 0.76 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr11_-_17981421 | 0.75 |
ENSDART00000005999
ENSDART00000104046 |
twf2b
|
twinfilin actin-binding protein 2b |
| chr15_-_15357178 | 0.75 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr7_+_29954709 | 0.73 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr3_+_59851537 | 0.72 |
ENSDART00000180997
|
CU693479.1
|
|
| chr1_-_614583 | 0.71 |
ENSDART00000004062
ENSDART00000147937 ENSDART00000146351 ENSDART00000134456 |
atp5pf
|
ATP synthase peripheral stalk subunit F6 |
| chr6_+_25257728 | 0.71 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr12_+_27127139 | 0.70 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr24_+_21622373 | 0.70 |
ENSDART00000183611
|
rpl21
|
ribosomal protein L21 |
| chr15_+_32711663 | 0.69 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr1_+_47459723 | 0.69 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr13_-_36535128 | 0.69 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr12_+_48216662 | 0.68 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr9_+_53337974 | 0.68 |
ENSDART00000145138
|
dct
|
dopachrome tautomerase |
| chr5_-_64831207 | 0.67 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr2_-_32356539 | 0.67 |
ENSDART00000169316
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr14_-_498979 | 0.66 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr21_-_26071773 | 0.66 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr15_+_28685625 | 0.65 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr4_+_14660769 | 0.64 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr20_+_15015557 | 0.64 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr4_+_2252123 | 0.64 |
ENSDART00000163996
ENSDART00000066491 |
glipr1a
|
GLI pathogenesis-related 1a |
| chr9_-_48370645 | 0.63 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr9_-_18911608 | 0.62 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr17_+_41992054 | 0.62 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr9_-_32753535 | 0.62 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr24_-_6078222 | 0.62 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr6_-_57655299 | 0.62 |
ENSDART00000083807
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr11_+_30729745 | 0.61 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_-_39488482 | 0.61 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr22_+_3153876 | 0.60 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr12_+_47663419 | 0.60 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr3_-_36260102 | 0.60 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr21_+_25765734 | 0.59 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr21_+_26071874 | 0.59 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr2_+_54641644 | 0.59 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr3_+_26081343 | 0.59 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr25_+_5044780 | 0.58 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr5_+_12515402 | 0.58 |
ENSDART00000103278
|
zgc:171242
|
zgc:171242 |
| chr10_-_29903165 | 0.58 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr6_-_58764672 | 0.57 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr6_+_7123980 | 0.57 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr11_+_14199802 | 0.57 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr12_+_48220584 | 0.57 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr2_-_689047 | 0.57 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr13_-_4223955 | 0.57 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_+_40576679 | 0.56 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr1_-_21723329 | 0.56 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr5_-_63515210 | 0.56 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr8_-_2230128 | 0.56 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr6_-_13187168 | 0.56 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr4_+_76705830 | 0.56 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr7_-_26408472 | 0.56 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr23_-_5719453 | 0.55 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr20_-_29052391 | 0.55 |
ENSDART00000193482
ENSDART00000017090 |
thbs1b
|
thrombospondin 1b |
| chr7_+_15872357 | 0.55 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr19_+_43969363 | 0.54 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr12_+_39034225 | 0.54 |
ENSDART00000154064
|
si:dkey-239b22.2
|
si:dkey-239b22.2 |
| chr14_-_1998501 | 0.53 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr20_-_40370736 | 0.53 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr5_-_41494831 | 0.52 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_-_19316985 | 0.52 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr6_-_15757867 | 0.52 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| chr1_-_54063520 | 0.52 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr1_+_52929185 | 0.52 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr10_+_38610741 | 0.51 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr3_-_22212764 | 0.51 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr23_+_4689626 | 0.51 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr19_+_42609132 | 0.51 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr1_-_10647484 | 0.51 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr5_+_42280372 | 0.50 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr1_+_52392511 | 0.50 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr25_+_14507567 | 0.50 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr24_-_32408404 | 0.50 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr23_-_22650992 | 0.50 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr19_-_12145765 | 0.49 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr16_-_35952789 | 0.49 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr23_+_19813677 | 0.49 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr1_-_22678471 | 0.48 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr6_-_54180699 | 0.48 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr7_-_26087807 | 0.48 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr24_-_26302375 | 0.48 |
ENSDART00000130696
|
cops9
|
COP9 signalosome subunit 9 |
| chr21_-_5881344 | 0.47 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr14_+_35691889 | 0.47 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr1_-_17569793 | 0.47 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr3_-_12930217 | 0.47 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr23_+_43493408 | 0.46 |
ENSDART00000191441
|
znf341
|
zinc finger protein 341 |
| chr10_-_9410068 | 0.46 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr15_-_47865063 | 0.46 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr6_+_13779857 | 0.46 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr21_+_10828828 | 0.46 |
ENSDART00000136449
|
oacyl
|
O-acyltransferase like |
| chr2_-_10192459 | 0.45 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr20_+_38032143 | 0.45 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_54908895 | 0.45 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr7_+_59169081 | 0.44 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr2_-_1514001 | 0.44 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr20_-_19422496 | 0.44 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr4_+_31293362 | 0.44 |
ENSDART00000169781
|
si:dkey-57l19.1
|
si:dkey-57l19.1 |
| chr1_-_17570013 | 0.44 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr18_+_7345417 | 0.44 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr7_+_20471315 | 0.44 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr17_-_6730247 | 0.44 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr9_-_2594410 | 0.44 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr9_+_13120419 | 0.44 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr25_-_10564721 | 0.44 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr15_-_42206890 | 0.43 |
ENSDART00000015843
|
pax3b
|
paired box 3b |
| chr22_+_11857356 | 0.43 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr20_-_4738101 | 0.43 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr12_+_8168272 | 0.43 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr24_-_37680917 | 0.43 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr22_-_26005894 | 0.43 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr7_-_69636502 | 0.43 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr10_-_40968095 | 0.43 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr16_-_50175069 | 0.42 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr17_+_21919131 | 0.42 |
ENSDART00000048136
|
htra1a
|
HtrA serine peptidase 1a |
| chr11_-_45138857 | 0.42 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_-_17960228 | 0.42 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr21_+_22423286 | 0.42 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr4_-_22671469 | 0.42 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr23_+_409578 | 0.42 |
ENSDART00000008177
|
slc25a55b
|
solute carrier family 25, member 55b |
| chr25_-_29134654 | 0.41 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr1_+_25696798 | 0.41 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr25_-_22187397 | 0.41 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr16_+_7653905 | 0.41 |
ENSDART00000081431
|
popdc3
|
popeye domain containing 3 |
| chr4_-_9722568 | 0.41 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr7_-_30174882 | 0.41 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr21_+_26726936 | 0.41 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr19_+_10847118 | 0.41 |
ENSDART00000189784
|
apoa4a
|
apolipoprotein A-IV a |
| chr15_-_22074315 | 0.41 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr4_-_18954001 | 0.41 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr10_-_20669635 | 0.41 |
ENSDART00000131361
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr14_-_4076480 | 0.40 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr2_+_16160906 | 0.40 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr4_+_5796761 | 0.40 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr21_-_16113477 | 0.40 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr16_+_31921812 | 0.40 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr16_+_16968682 | 0.40 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr7_+_34290051 | 0.39 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr5_+_67448664 | 0.39 |
ENSDART00000105731
|
si:dkey-251i10.1
|
si:dkey-251i10.1 |
| chr17_+_29276544 | 0.39 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr16_-_17072440 | 0.39 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr7_+_41295974 | 0.39 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr8_-_13574764 | 0.38 |
ENSDART00000076561
|
B3GNT3 (1 of many)
|
si:ch211-126g16.10 |
| chr21_-_44104600 | 0.38 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr1_-_44157273 | 0.38 |
ENSDART00000166618
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr5_-_26093945 | 0.38 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr9_-_9348058 | 0.38 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr6_+_56141852 | 0.38 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr10_+_9410304 | 0.38 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr8_-_18537866 | 0.38 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr18_+_46382484 | 0.38 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr1_-_10647307 | 0.37 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr7_-_34192834 | 0.37 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr16_+_42772678 | 0.37 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr1_-_35924495 | 0.37 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr13_+_7442023 | 0.37 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr14_-_13048355 | 0.37 |
ENSDART00000166434
|
si:dkey-35h6.1
|
si:dkey-35h6.1 |
| chr6_+_34028532 | 0.37 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 0.9 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 0.8 | GO:0090190 | regulation of neurotransmitter uptake(GO:0051580) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 0.7 | GO:2000648 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.2 | 0.7 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 0.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.8 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 4.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.6 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.6 | GO:0072314 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.4 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.0 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 1.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 1.0 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.4 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 1.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.5 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 5.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.6 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.3 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.6 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.3 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.8 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.1 | 0.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.2 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 0.5 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.6 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.6 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.3 | GO:0009099 | leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.2 | GO:0046385 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.1 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:2000379 | positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.1 | 0.3 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.3 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0048008 | positive regulation of protein autophosphorylation(GO:0031954) platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.1 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.6 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.8 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 2.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.2 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.5 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.2 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.4 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 4.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.4 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 1.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.0 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.5 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 1.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 1.8 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.6 | GO:1990266 | granulocyte migration(GO:0097530) neutrophil migration(GO:1990266) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.9 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.1 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.5 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.3 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.3 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 2.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 0.8 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.9 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.5 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 0.8 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.6 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.4 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 6.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 2.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.2 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 0.3 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.2 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.3 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 5.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0005231 | excitatory extracellular ligand-gated ion channel activity(GO:0005231) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 5.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |