Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
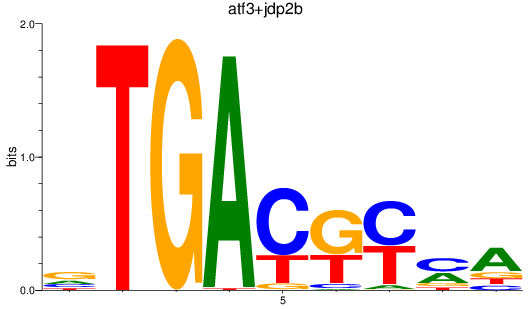
Results for atf3+jdp2b
Z-value: 2.54
Transcription factors associated with atf3+jdp2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf3
|
ENSDARG00000007823 | activating transcription factor 3 |
|
jdp2b
|
ENSDARG00000020133 | Jun dimerization protein 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jdp2b | dr11_v1_chr20_+_46586678_46586692 | -0.89 | 1.3e-03 | Click! |
| atf3 | dr11_v1_chr20_+_37794633_37794633 | 0.28 | 4.7e-01 | Click! |
Activity profile of atf3+jdp2b motif
Sorted Z-values of atf3+jdp2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_52442622 | 4.08 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr8_+_42917515 | 3.72 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr8_+_23725957 | 3.24 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr7_-_51773166 | 3.14 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr17_+_25187226 | 2.84 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr11_-_44801968 | 2.79 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr18_+_14645568 | 2.79 |
ENSDART00000138995
ENSDART00000147351 |
vps9d1
|
VPS9 domain containing 1 |
| chr5_-_3991655 | 2.72 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr10_+_6884123 | 2.70 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr22_+_17261801 | 2.70 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr8_+_52442785 | 2.60 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr4_-_16628801 | 2.58 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr22_+_24623936 | 2.57 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr2_-_55779927 | 2.53 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr1_-_52210950 | 2.37 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
| chr23_+_19590006 | 2.36 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr18_-_16953978 | 2.32 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr3_+_14463941 | 2.23 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr2_+_35603637 | 2.20 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr23_-_18030399 | 2.19 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr23_+_19790962 | 2.11 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr19_-_27570333 | 2.09 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr15_+_5116179 | 2.02 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr19_+_14454306 | 2.02 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr1_+_49955869 | 1.98 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr20_-_51831816 | 1.97 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr21_+_3928947 | 1.96 |
ENSDART00000149777
|
setx
|
senataxin |
| chr11_+_11120532 | 1.94 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr19_-_6840506 | 1.93 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr14_-_22495604 | 1.92 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr22_+_1170294 | 1.91 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr16_-_54919260 | 1.90 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr20_-_39789036 | 1.89 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr23_-_24226533 | 1.89 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr14_-_41468892 | 1.87 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr17_+_50657509 | 1.85 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr9_-_38021889 | 1.85 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr11_+_31680513 | 1.85 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr9_+_24126223 | 1.79 |
ENSDART00000132045
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr13_+_534453 | 1.78 |
ENSDART00000147909
|
wu:fc17b08
|
wu:fc17b08 |
| chr17_+_26722904 | 1.78 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr5_+_57658898 | 1.77 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr1_+_8521323 | 1.76 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr18_+_45526585 | 1.76 |
ENSDART00000138511
|
kifc3
|
kinesin family member C3 |
| chr5_+_26213874 | 1.75 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr8_-_5267442 | 1.75 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr5_+_13472234 | 1.75 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr12_-_21684197 | 1.74 |
ENSDART00000152999
ENSDART00000153109 ENSDART00000148698 |
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr18_+_14633974 | 1.73 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr14_+_30285613 | 1.72 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_-_24542952 | 1.72 |
ENSDART00000088777
|
atp13a2
|
ATPase 13A2 |
| chr5_-_29531948 | 1.71 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr8_+_50190742 | 1.71 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr7_-_69429561 | 1.71 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr3_-_49504023 | 1.70 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr12_-_27212880 | 1.68 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr7_-_20241346 | 1.67 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr8_+_48966165 | 1.66 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr2_-_17115256 | 1.66 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr15_-_24960730 | 1.65 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr12_-_27212596 | 1.65 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr15_-_41689981 | 1.64 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr9_-_34269066 | 1.64 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr5_-_69944084 | 1.60 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr14_-_763744 | 1.59 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr17_+_52612866 | 1.57 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr16_-_41535690 | 1.57 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr2_-_17114852 | 1.56 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_+_24597001 | 1.56 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr7_+_24153070 | 1.56 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr20_+_39223235 | 1.55 |
ENSDART00000132132
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr21_+_4256291 | 1.55 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr7_+_13491452 | 1.55 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr17_+_1544903 | 1.53 |
ENSDART00000156244
ENSDART00000112183 |
cep170b
|
centrosomal protein 170B |
| chr22_+_38935060 | 1.53 |
ENSDART00000183732
ENSDART00000130055 |
sirt7
|
sirtuin 7 |
| chr17_-_29312506 | 1.52 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr20_+_46572550 | 1.52 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr21_+_20901505 | 1.52 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr5_+_29831235 | 1.51 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr19_+_13994563 | 1.51 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr7_+_46019780 | 1.51 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr9_-_27749936 | 1.50 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr6_-_52484566 | 1.50 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr16_+_28547157 | 1.49 |
ENSDART00000109450
ENSDART00000165687 |
fam171a1
|
family with sequence similarity 171, member A1 |
| chr23_-_33738945 | 1.48 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr2_+_42871831 | 1.48 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr6_-_4228640 | 1.48 |
ENSDART00000162497
ENSDART00000179923 |
trak2
|
trafficking protein, kinesin binding 2 |
| chr24_-_40901410 | 1.48 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr19_+_43341424 | 1.47 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr11_-_13341483 | 1.47 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr16_-_36064143 | 1.47 |
ENSDART00000158358
ENSDART00000182584 |
stk40
|
serine/threonine kinase 40 |
| chr21_-_13784859 | 1.46 |
ENSDART00000024720
|
si:ch211-282j22.3
|
si:ch211-282j22.3 |
| chr4_-_17353972 | 1.46 |
ENSDART00000041529
|
parpbp
|
PARP1 binding protein |
| chr20_-_51831657 | 1.46 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr19_-_3781405 | 1.45 |
ENSDART00000170609
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr18_-_20444296 | 1.45 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr21_+_17051478 | 1.44 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr21_+_9997418 | 1.44 |
ENSDART00000181454
ENSDART00000171579 |
herc7
|
hect domain and RLD 7 |
| chr24_+_10898671 | 1.44 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr7_+_34549198 | 1.44 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr17_-_2690083 | 1.44 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr14_+_21755469 | 1.44 |
ENSDART00000186326
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr9_+_21793565 | 1.43 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr11_+_6295370 | 1.43 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr8_+_49117518 | 1.43 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr7_-_19999152 | 1.42 |
ENSDART00000173881
ENSDART00000100798 |
trip6
|
thyroid hormone receptor interactor 6 |
| chr17_-_31212420 | 1.42 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr5_-_5669879 | 1.42 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr19_-_3056235 | 1.42 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr17_+_32623931 | 1.42 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr8_-_410199 | 1.41 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr7_-_19998723 | 1.41 |
ENSDART00000173458
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr11_+_19433936 | 1.39 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr3_+_42923275 | 1.39 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr15_-_41689684 | 1.39 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr19_+_28291376 | 1.39 |
ENSDART00000139433
ENSDART00000103855 |
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr17_-_5610514 | 1.38 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr15_+_29140126 | 1.38 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr20_-_49961997 | 1.38 |
ENSDART00000187569
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr7_-_69352424 | 1.37 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr4_-_14915268 | 1.37 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr2_-_43739740 | 1.36 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr20_-_52939501 | 1.36 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_10563576 | 1.36 |
ENSDART00000185818
ENSDART00000190887 |
ccdc18
|
coiled-coil domain containing 18 |
| chr17_+_23554932 | 1.36 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr25_-_37084032 | 1.36 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr5_-_26247215 | 1.35 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr7_+_34549377 | 1.35 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr3_+_40255408 | 1.33 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr12_-_24832297 | 1.33 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr8_-_410728 | 1.33 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr17_-_6954719 | 1.32 |
ENSDART00000188180
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr21_+_20391978 | 1.32 |
ENSDART00000180817
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr10_+_14488625 | 1.32 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr17_+_30894431 | 1.31 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr17_+_28005763 | 1.31 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr2_-_10564019 | 1.30 |
ENSDART00000132167
|
ccdc18
|
coiled-coil domain containing 18 |
| chr10_+_16165533 | 1.30 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr2_-_57941037 | 1.29 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr11_+_42765963 | 1.29 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr19_+_43341115 | 1.29 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
| chr14_+_30413758 | 1.29 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr2_+_36121373 | 1.27 |
ENSDART00000187002
|
CT867973.2
|
|
| chr10_+_15603082 | 1.27 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr22_+_3184500 | 1.27 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr8_-_19266325 | 1.26 |
ENSDART00000036148
ENSDART00000137994 |
zgc:77486
|
zgc:77486 |
| chr14_+_8947282 | 1.26 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr19_-_25081711 | 1.26 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr21_+_45502773 | 1.25 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr15_+_35933094 | 1.25 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr2_-_37478418 | 1.25 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr2_-_32237916 | 1.25 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr25_+_36325793 | 1.25 |
ENSDART00000186973
|
HIST1H4E
|
zgc:165555 |
| chr5_-_16425781 | 1.24 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr5_-_18961694 | 1.24 |
ENSDART00000142531
ENSDART00000090521 |
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr15_+_39977461 | 1.24 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr22_+_12162470 | 1.24 |
ENSDART00000053045
ENSDART00000143599 |
ccnt2b
|
cyclin T2b |
| chr14_+_30413312 | 1.23 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr21_+_3775551 | 1.22 |
ENSDART00000111699
|
tor1
|
torsin family 1 |
| chr8_+_44714336 | 1.22 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr17_+_28102487 | 1.22 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr13_+_35856463 | 1.21 |
ENSDART00000171056
ENSDART00000017202 |
kcnk1b
|
potassium channel, subfamily K, member 1b |
| chr2_+_10821579 | 1.21 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr20_-_32981053 | 1.21 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr10_-_41285235 | 1.20 |
ENSDART00000141190
|
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr11_+_45153104 | 1.20 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr2_-_20866758 | 1.20 |
ENSDART00000165374
|
tpra
|
translocated promoter region a, nuclear basket protein |
| chr17_+_691453 | 1.19 |
ENSDART00000159271
|
fancm
|
Fanconi anemia, complementation group M |
| chr13_-_44782462 | 1.19 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr11_+_25328199 | 1.19 |
ENSDART00000141478
ENSDART00000112209 |
fam83d
|
family with sequence similarity 83, member D |
| chr5_-_48664522 | 1.19 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr19_+_27858866 | 1.19 |
ENSDART00000140336
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr20_-_211920 | 1.19 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr2_-_6292510 | 1.19 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr17_+_15674052 | 1.19 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr15_+_17100697 | 1.19 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr3_-_40933415 | 1.19 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr8_+_21254192 | 1.19 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr20_-_36671660 | 1.18 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr4_-_26035770 | 1.18 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr11_-_34783938 | 1.18 |
ENSDART00000135725
ENSDART00000039847 |
chchd4a
|
coiled-coil-helix-coiled-coil-helix domain containing 4a |
| chr13_+_47623916 | 1.18 |
ENSDART00000109266
|
mertka
|
c-mer proto-oncogene tyrosine kinase a |
| chr2_-_37280617 | 1.17 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr5_-_69523816 | 1.17 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr24_+_37461457 | 1.16 |
ENSDART00000165775
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr8_-_35960987 | 1.16 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr17_-_8592824 | 1.16 |
ENSDART00000127022
|
CU462878.1
|
|
| chr13_-_17464654 | 1.16 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr25_-_37071036 | 1.16 |
ENSDART00000150178
|
wwox
|
WW domain containing oxidoreductase |
| chr24_-_19718077 | 1.16 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr1_-_17693273 | 1.16 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr18_+_8917766 | 1.15 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr23_-_17004783 | 1.15 |
ENSDART00000126841
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr1_-_59240975 | 1.15 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr25_-_35113891 | 1.14 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr21_-_4695583 | 1.14 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr14_+_31618982 | 1.13 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr13_-_17464362 | 1.13 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr13_+_51710725 | 1.13 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr18_+_30441740 | 1.12 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf3+jdp2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.9 | 2.6 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.8 | 3.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.7 | 2.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.7 | 0.7 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.6 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 1.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.6 | 2.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.5 | 2.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.5 | 1.5 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.5 | 5.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.5 | 1.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 2.3 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.5 | 1.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 1.4 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.4 | 2.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.4 | 1.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.4 | 1.2 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.4 | 1.2 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.4 | 2.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 2.8 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.4 | 1.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 1.5 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.4 | 1.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.4 | 1.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.4 | 1.1 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.4 | 2.8 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.4 | 1.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.4 | 2.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.3 | 6.8 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.3 | 1.2 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 2.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.3 | 1.2 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 1.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 1.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 1.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 0.9 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.3 | 2.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 2.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 3.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.3 | 2.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 0.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 2.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.3 | 1.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 0.8 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 0.8 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 1.3 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 2.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 1.5 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.2 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.2 | 1.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.7 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 2.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 0.9 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 1.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.7 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 2.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.6 | GO:0044784 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) metaphase/anaphase transition of cell cycle(GO:0044784) |
| 0.2 | 1.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 0.9 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.7 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.2 | 2.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.2 | 1.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 1.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 1.0 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.6 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 2.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.8 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 3.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.2 | 1.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 0.5 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 0.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 1.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.5 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 0.7 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 1.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 1.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 1.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.8 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 0.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 1.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 1.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.2 | 0.6 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.2 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 0.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 0.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.5 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.5 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.8 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.1 | 2.0 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 1.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 4.7 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 2.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.9 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.6 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.8 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.1 | 0.4 | GO:0051306 | mitotic sister chromatid separation(GO:0051306) |
| 0.1 | 1.4 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.4 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 1.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 6.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 1.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 2.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.1 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.1 | 0.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 2.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.5 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 3.2 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 2.0 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.9 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.9 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 1.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.5 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.3 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 2.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.4 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.5 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.3 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 2.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.7 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.7 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.9 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 1.1 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.2 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 1.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.7 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.9 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 3.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 1.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 2.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 3.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.6 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 1.2 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 1.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.9 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 1.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 1.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 1.4 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 1.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of protein tyrosine kinase activity(GO:0061098) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 1.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 2.7 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.1 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.9 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.4 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 1.0 | GO:0006458 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.9 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.9 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.0 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 2.2 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 3.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 1.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 4.2 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 1.2 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.1 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 2.7 | GO:0030258 | lipid modification(GO:0030258) |
| 0.0 | 0.8 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 2.7 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 2.3 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.5 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.6 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.9 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 1.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.8 | 3.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 6.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 1.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 2.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 2.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 0.8 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 1.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.3 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 0.8 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.5 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 0.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.4 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 1.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 0.6 | GO:1990077 | primosome complex(GO:1990077) |
| 0.2 | 0.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 1.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 1.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 1.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 3.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 3.0 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 3.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 5.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 3.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 5.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 2.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 2.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 3.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 4.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.8 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 10.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.8 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.7 | 2.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.6 | 3.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.6 | 3.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 1.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.5 | 1.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 1.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.4 | 6.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 1.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.4 | 1.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.4 | 2.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.4 | 2.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 2.7 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.4 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 1.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.4 | 1.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.3 | 1.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 1.0 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.3 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.0 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 2.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 0.9 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 2.8 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.3 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 0.9 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.3 | 1.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 1.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 0.8 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 1.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 0.8 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 0.8 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 0.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 0.8 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.3 | 0.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 0.7 | GO:1901611 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.2 | 0.7 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 1.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.9 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.9 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 2.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 0.8 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 2.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.9 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.2 | 0.9 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 1.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.5 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 2.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 3.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 2.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 1.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 2.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 3.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 0.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 1.5 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.9 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 3.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 5.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.6 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.7 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 1.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 2.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.5 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 6.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 2.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.4 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 2.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 1.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 1.8 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 3.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.9 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 1.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 6.6 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.5 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 1.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 3.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.0 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 2.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 3.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.5 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 1.5 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.4 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 0.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 1.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 1.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 2.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |