Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
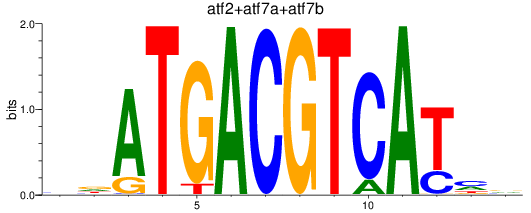
Results for atf2+atf7a+atf7b_creb5a+creb5b
Z-value: 0.51
Transcription factors associated with atf2+atf7a+atf7b_creb5a+creb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf7a
|
ENSDARG00000011298 | activating transcription factor 7a |
|
atf2
|
ENSDARG00000023903 | activating transcription factor 2 |
|
atf7b
|
ENSDARG00000055481 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000114492 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000115171 | activating transcription factor 7b |
|
creb5b
|
ENSDARG00000070536 | cAMP responsive element binding protein 5b |
|
creb5a
|
ENSDARG00000099002 | cAMP responsive element binding protein 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb5b | dr11_v1_chr16_-_20707742_20707742 | 0.88 | 1.6e-03 | Click! |
| creb5a | dr11_v1_chr19_-_19505167_19505167 | -0.24 | 5.3e-01 | Click! |
| atf2 | dr11_v1_chr9_+_2343096_2343172 | 0.07 | 8.7e-01 | Click! |
| atf7b | dr11_v1_chr6_-_39631164_39631181 | -0.01 | 9.8e-01 | Click! |
| atf7a | dr11_v1_chr23_-_27479558_27479558 | 0.01 | 9.9e-01 | Click! |
Activity profile of atf2+atf7a+atf7b_creb5a+creb5b motif
Sorted Z-values of atf2+atf7a+atf7b_creb5a+creb5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43606502 | 0.78 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr16_+_34523515 | 0.72 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr5_-_41494831 | 0.68 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr19_-_7225060 | 0.67 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr18_-_5598958 | 0.66 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr9_+_7548533 | 0.60 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr24_-_7632187 | 0.57 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr9_-_296169 | 0.54 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr18_-_7143920 | 0.52 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr3_-_49566364 | 0.51 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr25_-_32751982 | 0.47 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr1_-_22861348 | 0.45 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr20_+_29743904 | 0.43 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr16_-_12173554 | 0.41 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr6_+_60055168 | 0.41 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr16_+_5184402 | 0.41 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr24_+_7631797 | 0.41 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr3_-_50443607 | 0.40 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr20_-_32446406 | 0.40 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr20_+_20637866 | 0.39 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr1_-_38756870 | 0.39 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr15_+_28685892 | 0.39 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr21_+_45816030 | 0.39 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr5_+_56268436 | 0.39 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr15_+_28685625 | 0.36 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr7_+_40228422 | 0.35 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr4_-_69189894 | 0.35 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr20_+_20638034 | 0.35 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr25_+_19201231 | 0.35 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr19_-_1961024 | 0.34 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_+_69808763 | 0.34 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_-_42415902 | 0.34 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr13_+_36146415 | 0.33 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr12_+_5129245 | 0.33 |
ENSDART00000169073
|
pde6c
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr8_+_15239549 | 0.33 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr6_-_48311 | 0.33 |
ENSDART00000131010
|
zgc:114175
|
zgc:114175 |
| chr20_-_24122881 | 0.33 |
ENSDART00000131857
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr22_+_1796057 | 0.32 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr22_-_15593824 | 0.31 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr3_+_20156956 | 0.31 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr5_-_23999777 | 0.30 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr17_-_15382704 | 0.30 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr19_+_9174166 | 0.30 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr8_-_9118958 | 0.29 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr2_-_30324610 | 0.29 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr1_+_50968908 | 0.29 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr22_-_38621438 | 0.28 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr10_-_24371312 | 0.28 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr17_-_26911852 | 0.28 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr7_-_35516251 | 0.27 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr25_-_774350 | 0.26 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr14_-_11430566 | 0.26 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr17_+_15388479 | 0.26 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr8_+_39607466 | 0.25 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr16_-_16212615 | 0.25 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr24_-_26328721 | 0.25 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr14_+_790166 | 0.25 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr12_+_48634927 | 0.24 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr24_-_24163201 | 0.24 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr1_-_54947592 | 0.23 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr16_-_54942532 | 0.23 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr4_-_16853464 | 0.23 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr24_-_24162930 | 0.23 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr11_+_25638172 | 0.23 |
ENSDART00000114226
ENSDART00000143677 |
grm6b
|
glutamate receptor, metabotropic 6b |
| chr2_-_30324297 | 0.22 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr10_+_37145007 | 0.22 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr14_+_16813816 | 0.22 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr16_-_32649929 | 0.22 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr6_+_27667359 | 0.22 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr6_+_58832155 | 0.21 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr17_+_8184649 | 0.21 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr6_+_21227621 | 0.21 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr16_-_33097398 | 0.21 |
ENSDART00000166617
|
dopey1
|
dopey family member 1 |
| chr4_-_149334 | 0.21 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr16_+_24971717 | 0.20 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr22_+_997838 | 0.20 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr9_+_219124 | 0.20 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr4_-_73787702 | 0.20 |
ENSDART00000136328
ENSDART00000150546 |
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr5_+_9047433 | 0.20 |
ENSDART00000091463
|
idua
|
iduronidase, alpha-L- |
| chr24_+_4373355 | 0.19 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr1_+_51475094 | 0.19 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr23_+_45456490 | 0.19 |
ENSDART00000036631
|
cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr10_-_20523405 | 0.19 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr24_+_4373033 | 0.19 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr6_+_13920479 | 0.19 |
ENSDART00000155480
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr14_+_11950011 | 0.19 |
ENSDART00000188138
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr11_+_575665 | 0.18 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr14_-_34771864 | 0.18 |
ENSDART00000141157
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr10_+_6496185 | 0.18 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr7_-_27686021 | 0.18 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr13_-_31470439 | 0.18 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr24_+_744713 | 0.18 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr5_+_45322734 | 0.17 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr11_-_44876005 | 0.17 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr25_+_35683956 | 0.17 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr3_-_5964557 | 0.17 |
ENSDART00000184738
|
BX284638.1
|
|
| chr20_-_9462433 | 0.17 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr18_+_910992 | 0.16 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr15_+_1004680 | 0.16 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr8_-_65189 | 0.16 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr18_+_46773198 | 0.16 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr10_-_43117607 | 0.16 |
ENSDART00000148293
ENSDART00000089965 |
tmem167a
|
transmembrane protein 167A |
| chr13_-_50672341 | 0.16 |
ENSDART00000190498
|
FP102122.1
|
|
| chr4_-_6373735 | 0.16 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr6_-_14010554 | 0.15 |
ENSDART00000004656
|
zgc:92027
|
zgc:92027 |
| chr9_+_54178475 | 0.15 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr21_-_23308286 | 0.15 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr6_+_58832323 | 0.15 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr18_-_19103929 | 0.15 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr10_-_9089545 | 0.15 |
ENSDART00000080781
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr18_+_12058403 | 0.14 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr7_+_23907692 | 0.14 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr7_+_22616212 | 0.14 |
ENSDART00000052844
|
cldn7a
|
claudin 7a |
| chr17_-_13007484 | 0.14 |
ENSDART00000156812
|
si:dkeyp-33b5.4
|
si:dkeyp-33b5.4 |
| chr4_-_72544805 | 0.13 |
ENSDART00000110261
|
zgc:175107
|
zgc:175107 |
| chr10_-_42882215 | 0.13 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr12_-_36740781 | 0.13 |
ENSDART00000105484
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr11_-_44163164 | 0.13 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr10_+_44903676 | 0.13 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr20_-_34801181 | 0.13 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr7_-_12065668 | 0.13 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr17_+_22956427 | 0.13 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr18_+_17663898 | 0.13 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr5_-_24000211 | 0.13 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr19_-_26923273 | 0.13 |
ENSDART00000089609
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr11_+_42474694 | 0.13 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr3_-_2593859 | 0.13 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr4_+_40953320 | 0.13 |
ENSDART00000151912
|
znf1136
|
zinc finger protein 1136 |
| chr12_+_48220584 | 0.13 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr23_-_4915118 | 0.12 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr17_+_29345606 | 0.12 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr9_-_33121535 | 0.12 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr15_-_12319065 | 0.12 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr20_-_18915376 | 0.12 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr4_-_73825089 | 0.12 |
ENSDART00000174207
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr8_+_21588067 | 0.12 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr22_+_30330574 | 0.12 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr25_-_7686201 | 0.12 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr17_-_24866964 | 0.12 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr14_+_597532 | 0.12 |
ENSDART00000159805
|
LO018568.2
|
|
| chr16_+_46111849 | 0.12 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_+_2279051 | 0.12 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr5_+_51848756 | 0.11 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
| chr25_+_28893615 | 0.11 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr13_+_23843712 | 0.11 |
ENSDART00000057611
|
oprm1
|
opioid receptor, mu 1 |
| chr11_+_329687 | 0.11 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr4_+_77151419 | 0.11 |
ENSDART00000174312
|
CU467646.4
|
|
| chr18_-_46063773 | 0.11 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr13_+_32446169 | 0.11 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr5_-_20814576 | 0.11 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr4_+_30775376 | 0.10 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr4_+_72578548 | 0.10 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr17_+_12698532 | 0.10 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr4_-_55641422 | 0.10 |
ENSDART00000165178
|
znf1074
|
zinc finger protein 1074 |
| chr13_-_40499296 | 0.10 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr14_+_11430796 | 0.10 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr21_-_3770636 | 0.10 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr11_+_42478184 | 0.10 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr15_+_5028608 | 0.10 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr2_+_22694382 | 0.10 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr23_-_24146591 | 0.09 |
ENSDART00000133269
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr8_-_24113575 | 0.09 |
ENSDART00000099692
ENSDART00000186211 |
dclre1b
|
DNA cross-link repair 1B |
| chr17_-_43666166 | 0.09 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr22_+_35472653 | 0.09 |
ENSDART00000076424
ENSDART00000187204 |
tctex1d2
|
Tctex1 domain containing 2 |
| chr21_-_5007109 | 0.09 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr8_-_23194408 | 0.09 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr21_-_25395223 | 0.09 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr25_-_26893006 | 0.09 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr12_-_5455936 | 0.09 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr15_-_28200049 | 0.09 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr12_-_17201028 | 0.09 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr1_-_44037843 | 0.09 |
ENSDART00000160276
|
si:ch73-109d9.3
|
si:ch73-109d9.3 |
| chr7_+_29301497 | 0.09 |
ENSDART00000099327
|
rab8b
|
RAB8B, member RAS oncogene family |
| chr15_-_27710513 | 0.09 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr4_-_1839352 | 0.08 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr6_-_11614339 | 0.08 |
ENSDART00000080589
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr23_-_4409668 | 0.08 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr25_+_16945348 | 0.08 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr15_+_618081 | 0.08 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr21_+_9576176 | 0.08 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr14_-_48939560 | 0.08 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr23_-_5759242 | 0.08 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr2_-_20994126 | 0.08 |
ENSDART00000062573
|
nrn1b
|
neuritin 1b |
| chr4_-_22363709 | 0.08 |
ENSDART00000037670
|
orc5
|
origin recognition complex, subunit 5 |
| chr7_+_48761875 | 0.08 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr7_-_34927961 | 0.08 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T cells 3a |
| chr10_-_17170086 | 0.08 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr21_+_30351256 | 0.08 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr4_+_76403698 | 0.08 |
ENSDART00000184821
ENSDART00000169373 |
FP074874.2
FP074874.1
|
|
| chr3_+_32403758 | 0.08 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr21_+_22845317 | 0.08 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr16_-_20707742 | 0.08 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr7_-_12909352 | 0.08 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr2_+_20332044 | 0.08 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr1_-_9940494 | 0.07 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr15_-_21692630 | 0.07 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr5_-_28559870 | 0.07 |
ENSDART00000154495
|
FP015972.1
|
|
| chr18_+_507835 | 0.07 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr15_+_1796313 | 0.07 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr14_-_48103207 | 0.07 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf2+atf7a+atf7b_creb5a+creb5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.6 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0090189 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.2 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.3 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |