Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
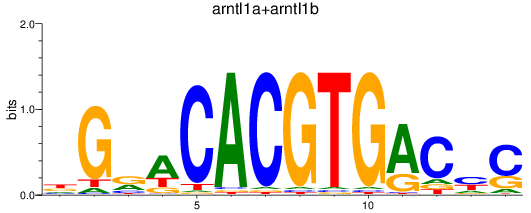
Results for arntl1a+arntl1b
Z-value: 0.79
Transcription factors associated with arntl1a+arntl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arntl1a
|
ENSDARG00000006791 | aryl hydrocarbon receptor nuclear translocator-like 1a |
|
arntl1b
|
ENSDARG00000035732 | aryl hydrocarbon receptor nuclear translocator-like 1b |
|
arntl1b
|
ENSDARG00000114562 | aryl hydrocarbon receptor nuclear translocator-like 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arntl1a | dr11_v1_chr25_-_17918810_17918810 | -0.87 | 2.2e-03 | Click! |
| arntl1b | dr11_v1_chr7_+_66072580_66072580 | -0.45 | 2.3e-01 | Click! |
Activity profile of arntl1a+arntl1b motif
Sorted Z-values of arntl1a+arntl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_12968101 | 1.78 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr24_-_12938922 | 1.66 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr24_-_33756003 | 1.22 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr9_+_40825065 | 1.19 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr24_+_35564668 | 1.15 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr7_+_55633483 | 1.05 |
ENSDART00000180993
ENSDART00000184845 |
trappc2l
|
trafficking protein particle complex 2-like |
| chr1_+_54766943 | 1.03 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr6_-_37422841 | 0.93 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr10_-_6775271 | 0.91 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr6_+_56147812 | 0.88 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr25_+_245438 | 0.82 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr6_-_32093830 | 0.79 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr21_-_4539899 | 0.78 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr22_-_94352 | 0.77 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr20_+_46213553 | 0.74 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr7_+_38750871 | 0.73 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr23_-_5719453 | 0.72 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr23_+_39606108 | 0.72 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr23_+_36095260 | 0.71 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr7_+_48999723 | 0.70 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr16_+_6864608 | 0.70 |
ENSDART00000078306
|
arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr4_+_842010 | 0.70 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr15_-_1198886 | 0.69 |
ENSDART00000063285
|
lxn
|
latexin |
| chr9_-_44295071 | 0.69 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr6_+_45918981 | 0.68 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr4_-_13921185 | 0.68 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr22_+_10646928 | 0.68 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr19_-_22346582 | 0.66 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr14_+_25465346 | 0.65 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr19_+_19750101 | 0.64 |
ENSDART00000168041
ENSDART00000170697 |
hoxa9a
CR382300.2
|
homeobox A9a |
| chr15_+_1148074 | 0.63 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr1_-_18803919 | 0.62 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr3_+_23737795 | 0.62 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr8_+_19674369 | 0.61 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr7_-_30082931 | 0.60 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr7_+_7048245 | 0.60 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr16_+_20915319 | 0.59 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr2_+_10280645 | 0.59 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr25_+_22107643 | 0.59 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr19_+_19737214 | 0.58 |
ENSDART00000160283
ENSDART00000169017 |
hoxa11a
|
homeobox A11a |
| chr25_+_37480285 | 0.58 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr10_-_22803740 | 0.54 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr9_-_24413008 | 0.54 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr24_+_7631797 | 0.53 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr13_-_31441042 | 0.50 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr19_-_32518556 | 0.50 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr3_+_23768898 | 0.50 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr25_-_6223567 | 0.50 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr4_+_7876197 | 0.50 |
ENSDART00000111986
ENSDART00000189601 |
cdc123
|
cell division cycle 123 homolog (S. cerevisiae) |
| chr24_+_39105051 | 0.49 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr2_+_50967947 | 0.49 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr7_+_6969909 | 0.49 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr19_+_19772765 | 0.49 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr10_-_44027391 | 0.48 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr5_-_10002260 | 0.48 |
ENSDART00000141831
|
si:ch73-266o15.4
|
si:ch73-266o15.4 |
| chr1_+_56447107 | 0.48 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr23_+_26079467 | 0.48 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr13_+_29510023 | 0.47 |
ENSDART00000187398
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr19_-_791016 | 0.47 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr6_+_3816241 | 0.47 |
ENSDART00000178545
|
erich2
|
glutamate-rich 2 |
| chr1_+_29178117 | 0.46 |
ENSDART00000114536
|
si:ch211-198k9.6
|
si:ch211-198k9.6 |
| chr24_-_982443 | 0.46 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr21_-_20765338 | 0.46 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr8_+_34922981 | 0.46 |
ENSDART00000188445
ENSDART00000188818 ENSDART00000182965 ENSDART00000180718 |
BX005361.1
|
|
| chr9_-_24242592 | 0.46 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr11_+_42556395 | 0.45 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr3_+_23752150 | 0.45 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr18_+_17428506 | 0.45 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr1_+_7956030 | 0.45 |
ENSDART00000159655
|
CR855320.1
|
|
| chr24_-_18919562 | 0.45 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr5_+_40224938 | 0.44 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr1_-_35916247 | 0.43 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr9_+_907459 | 0.42 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr8_+_34624025 | 0.42 |
ENSDART00000180772
|
BX247865.2
|
|
| chr7_-_40993456 | 0.40 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr11_+_14199802 | 0.40 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr13_+_281214 | 0.40 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr24_-_42090635 | 0.39 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr8_+_34439052 | 0.39 |
ENSDART00000193021
ENSDART00000193851 ENSDART00000183170 ENSDART00000181227 |
zgc:174461
|
zgc:174461 |
| chr1_+_45754868 | 0.39 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr9_-_27805801 | 0.39 |
ENSDART00000140608
ENSDART00000114542 |
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr19_-_26736336 | 0.39 |
ENSDART00000109258
ENSDART00000182802 |
csnk2b
|
casein kinase 2, beta polypeptide |
| chr8_+_34434345 | 0.39 |
ENSDART00000190246
ENSDART00000189447 ENSDART00000185557 ENSDART00000189230 |
zgc:174461
|
zgc:174461 |
| chr8_+_46386601 | 0.38 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr8_+_34443759 | 0.38 |
ENSDART00000187788
ENSDART00000187679 ENSDART00000188872 ENSDART00000186358 |
zgc:174461
|
zgc:174461 |
| chr8_+_34720244 | 0.38 |
ENSDART00000181958
ENSDART00000189806 ENSDART00000190167 ENSDART00000183165 |
zgc:174461
|
zgc:174461 |
| chr25_-_19585010 | 0.38 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr18_+_17428258 | 0.38 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr12_-_49168398 | 0.38 |
ENSDART00000186608
|
FO704624.1
|
|
| chr14_-_25452503 | 0.37 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr8_+_35032633 | 0.37 |
ENSDART00000184683
ENSDART00000184109 |
zgc:77614
|
zgc:77614 |
| chr3_+_23738215 | 0.37 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr8_-_34767412 | 0.36 |
ENSDART00000164901
|
CABZ01049825.1
|
|
| chr24_+_39211288 | 0.36 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr8_+_34988481 | 0.36 |
ENSDART00000186808
ENSDART00000164942 ENSDART00000186472 ENSDART00000182717 |
zgc:174461
|
zgc:174461 |
| chr3_+_39568290 | 0.36 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr3_-_11008532 | 0.35 |
ENSDART00000165086
|
CR382337.3
|
|
| chr22_+_38159823 | 0.35 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr19_-_26823647 | 0.34 |
ENSDART00000002464
|
neu1
|
neuraminidase 1 |
| chr16_-_24612871 | 0.34 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr8_+_34567717 | 0.34 |
ENSDART00000141506
|
BX005399.1
|
|
| chr21_-_15674802 | 0.33 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr25_-_19584735 | 0.33 |
ENSDART00000137930
|
sycp3
|
synaptonemal complex protein 3 |
| chr4_-_2036620 | 0.33 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr8_+_34448496 | 0.33 |
ENSDART00000188072
ENSDART00000180224 ENSDART00000189510 ENSDART00000190390 |
zgc:174461
|
zgc:174461 |
| chr8_+_34932397 | 0.33 |
ENSDART00000193631
ENSDART00000188761 ENSDART00000181280 ENSDART00000191784 ENSDART00000187029 |
zgc:174461
|
zgc:174461 |
| chr24_+_3307857 | 0.32 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr8_+_34486656 | 0.32 |
ENSDART00000136887
ENSDART00000160081 ENSDART00000162349 ENSDART00000183007 ENSDART00000185735 |
zgc:77614
|
zgc:77614 |
| chr8_+_34531954 | 0.32 |
ENSDART00000184878
ENSDART00000192417 ENSDART00000184988 ENSDART00000192039 |
zgc:174461
|
zgc:174461 |
| chr8_+_34540801 | 0.32 |
ENSDART00000186390
ENSDART00000184590 ENSDART00000187379 ENSDART00000180922 ENSDART00000187547 |
zgc:174461
|
zgc:174461 |
| chr8_+_34562983 | 0.32 |
ENSDART00000179915
ENSDART00000182870 ENSDART00000191069 ENSDART00000193201 ENSDART00000187830 |
zgc:174461
|
zgc:174461 |
| chr8_+_34572359 | 0.32 |
ENSDART00000187498
ENSDART00000184035 ENSDART00000193944 ENSDART00000185472 ENSDART00000193433 |
zgc:174461
|
zgc:174461 |
| chr8_+_34685407 | 0.32 |
ENSDART00000111653
ENSDART00000186260 ENSDART00000190456 ENSDART00000184348 ENSDART00000180741 |
zgc:174461
|
zgc:174461 |
| chr8_+_34927665 | 0.32 |
ENSDART00000186136
ENSDART00000192243 ENSDART00000182256 ENSDART00000186057 |
zgc:174461
|
zgc:174461 |
| chr1_+_17695426 | 0.32 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr8_+_34629184 | 0.32 |
ENSDART00000188917
ENSDART00000180349 ENSDART00000191426 |
zgc:174461
|
zgc:174461 |
| chr20_-_2949028 | 0.32 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr8_+_34527239 | 0.32 |
ENSDART00000190604
ENSDART00000179883 ENSDART00000180594 ENSDART00000183138 ENSDART00000183360 |
zgc:174461
|
zgc:174461 |
| chr8_-_25235676 | 0.32 |
ENSDART00000062363
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr20_-_25631256 | 0.32 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr21_-_13672195 | 0.31 |
ENSDART00000146717
|
clic3
|
chloride intracellular channel 3 |
| chr4_+_17353714 | 0.31 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr24_-_36316104 | 0.31 |
ENSDART00000048046
|
naglu
|
N-acetylglucosaminidase, alpha |
| chr19_+_7567763 | 0.31 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr21_+_11415224 | 0.30 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr6_-_36552844 | 0.30 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr8_+_34476532 | 0.30 |
ENSDART00000181956
ENSDART00000191537 |
zgc:174461
|
zgc:174461 |
| chr8_+_34998570 | 0.30 |
ENSDART00000184294
|
zgc:77614
|
zgc:77614 |
| chr4_-_64709908 | 0.28 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr18_-_6881392 | 0.28 |
ENSDART00000154968
|
si:dkey-266m15.7
|
si:dkey-266m15.7 |
| chr21_-_42100471 | 0.28 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr5_-_43959972 | 0.28 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr19_+_15440841 | 0.28 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_+_37268900 | 0.28 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr22_-_1237003 | 0.28 |
ENSDART00000169746
|
ampd2a
|
adenosine monophosphate deaminase 2a |
| chr19_+_19762183 | 0.28 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr11_+_42482920 | 0.27 |
ENSDART00000160937
|
arf4a
|
ADP-ribosylation factor 4a |
| chr24_+_42074143 | 0.27 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr11_-_236984 | 0.27 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr19_+_19756425 | 0.27 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr7_+_34620418 | 0.27 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr3_+_15817644 | 0.27 |
ENSDART00000055787
|
natd1
|
zgc:110779 |
| chr24_-_16917086 | 0.26 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr22_-_11136625 | 0.26 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr8_+_24745041 | 0.26 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr18_+_44649804 | 0.26 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr7_-_13381129 | 0.26 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr4_-_17669881 | 0.25 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr3_-_18756076 | 0.25 |
ENSDART00000055766
|
zgc:113333
|
zgc:113333 |
| chr21_+_6780340 | 0.25 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr25_+_19095231 | 0.25 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr22_-_38360205 | 0.25 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr7_+_49654588 | 0.25 |
ENSDART00000025451
ENSDART00000141934 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr9_-_27771339 | 0.25 |
ENSDART00000135722
ENSDART00000140381 |
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr12_+_13652361 | 0.24 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr4_+_16725960 | 0.24 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr22_-_94519 | 0.24 |
ENSDART00000161432
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr21_-_45382112 | 0.24 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr2_+_33189582 | 0.24 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
| chr4_+_63769987 | 0.24 |
ENSDART00000168878
|
znf1048
|
zinc finger protein 1048 |
| chr7_-_67214972 | 0.24 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr6_-_18976168 | 0.24 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr2_-_51096647 | 0.23 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr20_+_32406011 | 0.23 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr9_-_27771182 | 0.23 |
ENSDART00000170931
|
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr5_-_61349059 | 0.23 |
ENSDART00000136553
|
si:ch211-209a2.2
|
si:ch211-209a2.2 |
| chr3_-_16055432 | 0.23 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr25_+_37443194 | 0.23 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr16_+_41015163 | 0.23 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr12_+_13652747 | 0.23 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr3_-_48993580 | 0.23 |
ENSDART00000182400
|
BX769173.1
|
|
| chr20_+_13969414 | 0.22 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr1_+_29178331 | 0.22 |
ENSDART00000186905
|
si:ch211-198k9.6
|
si:ch211-198k9.6 |
| chr14_+_94946 | 0.21 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr4_-_38033800 | 0.21 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr15_-_37850969 | 0.21 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr7_+_48761875 | 0.21 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr18_-_6943577 | 0.21 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr17_-_42492668 | 0.20 |
ENSDART00000183946
|
CR925755.2
|
|
| chr8_-_26961779 | 0.20 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr4_-_38476901 | 0.20 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr18_-_1414760 | 0.20 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr24_-_7632187 | 0.20 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr20_+_32118559 | 0.20 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr3_-_1190132 | 0.20 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr2_-_10386738 | 0.20 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr15_-_3094516 | 0.19 |
ENSDART00000179719
|
slitrk5a
|
SLIT and NTRK-like family, member 5a |
| chr18_+_26337869 | 0.19 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr25_-_35599887 | 0.19 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr23_+_13814978 | 0.18 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr9_-_27805644 | 0.18 |
ENSDART00000192431
|
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr8_-_17167819 | 0.18 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr6_-_10912424 | 0.18 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr5_-_55395964 | 0.18 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr9_-_41784799 | 0.17 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr16_+_26824691 | 0.17 |
ENSDART00000135053
|
zmp:0000001316
|
zmp:0000001316 |
| chr7_-_7493758 | 0.17 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr23_+_31913292 | 0.17 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr15_-_41290415 | 0.17 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr23_+_25135858 | 0.16 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of arntl1a+arntl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 2.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 2.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.2 | 0.8 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.9 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.5 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 0.9 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 2.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.5 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.5 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |