Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
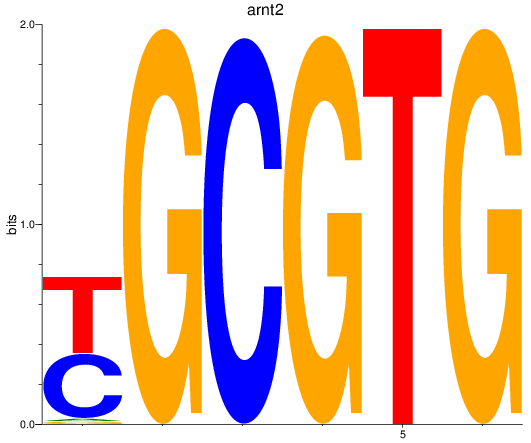
Results for arnt2
Z-value: 2.64
Transcription factors associated with arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arnt2
|
ENSDARG00000103697 | aryl-hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arnt2 | dr11_v1_chr7_+_10701770_10701770 | 0.44 | 2.3e-01 | Click! |
Activity profile of arnt2 motif
Sorted Z-values of arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_74090168 | 6.59 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr14_+_46313135 | 5.63 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr14_+_46313396 | 4.95 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr9_-_44295071 | 4.56 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr7_+_49715750 | 4.33 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr21_+_6751405 | 4.19 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr21_+_6751760 | 4.15 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr23_-_6641223 | 3.20 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr18_-_23875219 | 2.70 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_25326296 | 2.66 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr5_-_23429228 | 2.64 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr7_-_38340674 | 2.61 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr20_+_35382482 | 2.60 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_+_36932718 | 2.50 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr17_+_52822831 | 2.47 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr9_+_32978302 | 2.38 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr4_+_17417111 | 2.36 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr1_-_22757145 | 2.32 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr16_+_23978978 | 2.22 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr8_+_29593986 | 2.17 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr18_-_23875370 | 2.15 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr21_+_6780340 | 2.08 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr5_+_34997763 | 2.06 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr6_-_26559921 | 2.05 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr11_+_7324704 | 2.05 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr2_+_42724404 | 2.05 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr21_-_20711739 | 1.94 |
ENSDART00000190918
|
BX530077.1
|
|
| chr21_+_28958471 | 1.91 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr24_-_3419998 | 1.89 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr18_+_7345417 | 1.88 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr20_-_20533865 | 1.83 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr1_-_45633955 | 1.81 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr1_+_31942961 | 1.78 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr5_-_48285756 | 1.77 |
ENSDART00000183495
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr17_+_22956816 | 1.76 |
ENSDART00000079460
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_29710857 | 1.74 |
ENSDART00000021987
|
ptf1a
|
pancreas specific transcription factor, 1a |
| chr11_+_30513656 | 1.72 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr5_+_36415978 | 1.66 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr17_-_21784152 | 1.60 |
ENSDART00000127254
|
hmx2
|
H6 family homeobox 2 |
| chr16_+_19732543 | 1.60 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr3_+_23737795 | 1.58 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr15_+_33070939 | 1.57 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr18_-_23874929 | 1.55 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_-_11640240 | 1.53 |
ENSDART00000091752
|
fnbp1a
|
formin binding protein 1a |
| chr18_+_2837563 | 1.51 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr14_+_790166 | 1.47 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr17_+_52823015 | 1.47 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr12_+_27117609 | 1.46 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr23_-_31512496 | 1.45 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr21_-_41305748 | 1.45 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr1_+_36436936 | 1.45 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr25_+_13406069 | 1.42 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr14_+_33722950 | 1.39 |
ENSDART00000075312
|
apln
|
apelin |
| chr5_-_14344647 | 1.37 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr8_-_19904124 | 1.37 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr23_-_26077038 | 1.36 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr20_-_47731768 | 1.36 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr8_+_19674369 | 1.35 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr10_-_17484762 | 1.35 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr21_+_23953181 | 1.34 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr2_+_47581997 | 1.32 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_-_22756898 | 1.32 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr12_-_22540943 | 1.31 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr15_+_38859648 | 1.31 |
ENSDART00000141086
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr21_+_19445942 | 1.30 |
ENSDART00000030887
|
slc45a2
|
solute carrier family 45, member 2 |
| chr15_+_36156986 | 1.30 |
ENSDART00000059791
|
sst1.1
|
somatostatin 1, tandem duplicate 1 |
| chr17_-_42218652 | 1.30 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr19_-_47452874 | 1.28 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr21_+_17768174 | 1.27 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr14_+_36220479 | 1.26 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr19_+_17259912 | 1.26 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr10_-_26744131 | 1.24 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr23_+_22656477 | 1.24 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr11_-_24428237 | 1.23 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr14_+_8275115 | 1.22 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr4_-_4706893 | 1.21 |
ENSDART00000093005
|
CABZ01020835.1
|
|
| chr5_+_32162684 | 1.18 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr18_-_46208581 | 1.18 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr17_+_21919131 | 1.17 |
ENSDART00000048136
|
htra1a
|
HtrA serine peptidase 1a |
| chr6_+_2093206 | 1.16 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr19_+_30387999 | 1.16 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr21_+_5169154 | 1.16 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr9_+_25777270 | 1.15 |
ENSDART00000188547
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr19_+_30388186 | 1.15 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr23_-_8373676 | 1.15 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr17_+_22956427 | 1.14 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr19_+_48176745 | 1.14 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr9_+_13120419 | 1.14 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr6_+_2093367 | 1.12 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr13_-_40411908 | 1.12 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr9_-_23217196 | 1.12 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr4_-_27350820 | 1.12 |
ENSDART00000145806
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr19_-_10243148 | 1.12 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr18_-_44129151 | 1.11 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr21_+_17110598 | 1.09 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr23_-_31555696 | 1.08 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr20_+_34326874 | 1.08 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr5_+_23118470 | 1.07 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr12_+_35119762 | 1.06 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr11_+_36989696 | 1.05 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr23_+_10146542 | 1.04 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr16_+_10918252 | 1.04 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr14_-_17563773 | 1.04 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr24_-_8831866 | 1.04 |
ENSDART00000066780
ENSDART00000143501 |
gcm2
|
glial cells missing homolog 2 (Drosophila) |
| chr17_+_38262408 | 1.04 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr24_+_23742690 | 1.04 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr12_-_431249 | 1.04 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr19_+_49721 | 1.03 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr5_-_51619742 | 1.03 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr10_+_22012389 | 1.03 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr19_+_26718074 | 1.03 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr20_+_20637866 | 1.02 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr7_-_26924903 | 1.01 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr13_-_30027730 | 1.00 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr6_-_55997350 | 1.00 |
ENSDART00000011652
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr18_-_44610992 | 0.99 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr19_+_15571290 | 0.99 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr23_-_44494401 | 0.99 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr13_+_27314795 | 0.98 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr15_-_18574716 | 0.96 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr16_+_46294337 | 0.96 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr4_-_2867461 | 0.95 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr14_-_7885707 | 0.94 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr9_-_6661657 | 0.94 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr17_+_21964472 | 0.94 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr18_+_49411417 | 0.94 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr9_-_1978090 | 0.93 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr6_+_42818963 | 0.92 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr15_-_12319065 | 0.92 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr4_-_22311610 | 0.92 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr14_+_16813816 | 0.91 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr6_+_4872883 | 0.90 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr24_+_23791758 | 0.89 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr4_-_12007404 | 0.89 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr7_+_20017211 | 0.89 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr18_+_5490668 | 0.88 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr13_+_12299997 | 0.88 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr4_+_7888047 | 0.88 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr10_+_36650222 | 0.87 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr16_-_33059246 | 0.87 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr3_+_23726148 | 0.87 |
ENSDART00000174580
|
hoxb3a
|
homeobox B3a |
| chr5_+_48683447 | 0.87 |
ENSDART00000008043
|
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr20_-_8419057 | 0.86 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_-_3166726 | 0.86 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr17_-_22067451 | 0.85 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr10_-_15919839 | 0.85 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr20_+_826459 | 0.85 |
ENSDART00000104740
|
nt5e
|
5'-nucleotidase, ecto (CD73) |
| chr5_+_30179010 | 0.85 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr1_+_33383971 | 0.85 |
ENSDART00000150043
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr5_-_16983336 | 0.84 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr4_-_17055782 | 0.84 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr6_-_32703317 | 0.84 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr9_-_34945566 | 0.84 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr19_-_10395683 | 0.84 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr13_+_12739283 | 0.83 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr7_+_55633483 | 0.83 |
ENSDART00000180993
ENSDART00000184845 |
trappc2l
|
trafficking protein particle complex 2-like |
| chr6_-_18698006 | 0.82 |
ENSDART00000170128
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr24_+_26932472 | 0.82 |
ENSDART00000192288
ENSDART00000079848 |
tnfsf10
|
TNF superfamily member 10 |
| chr14_-_26704829 | 0.82 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr23_+_31107685 | 0.82 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr3_-_26109322 | 0.82 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr3_+_14768364 | 0.81 |
ENSDART00000090235
ENSDART00000139001 |
nfixb
|
nuclear factor I/Xb |
| chr13_+_31603988 | 0.81 |
ENSDART00000030646
|
six6a
|
SIX homeobox 6a |
| chr17_-_21441464 | 0.80 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr1_+_37195465 | 0.80 |
ENSDART00000043855
ENSDART00000192580 ENSDART00000181666 |
dclk2a
|
doublecortin-like kinase 2a |
| chr20_-_6812688 | 0.80 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr3_+_18097700 | 0.79 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr5_-_22952156 | 0.78 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr5_-_38121612 | 0.78 |
ENSDART00000159543
|
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr3_-_25813426 | 0.78 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr17_+_27434626 | 0.78 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr19_-_5369486 | 0.78 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr19_+_41464870 | 0.78 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr7_+_25858380 | 0.77 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr20_-_29505863 | 0.76 |
ENSDART00000148278
|
klf11a
|
Kruppel-like factor 11a |
| chr13_-_31296358 | 0.76 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr15_+_28482862 | 0.75 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr17_+_17861681 | 0.75 |
ENSDART00000123311
|
ism2a
|
isthmin 2a |
| chr4_-_25181552 | 0.75 |
ENSDART00000066930
|
atp5f1c
|
ATP synthase F1 subunit gamma |
| chr23_+_17220986 | 0.74 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr21_-_25522510 | 0.74 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr1_+_33969015 | 0.73 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr1_-_22512063 | 0.72 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr2_+_6991208 | 0.72 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr3_-_3496738 | 0.71 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr6_-_11614339 | 0.71 |
ENSDART00000080589
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr6_-_7720332 | 0.71 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr1_-_49434798 | 0.71 |
ENSDART00000150842
ENSDART00000150789 |
ATP5MD
|
si:dkeyp-80c12.10 |
| chr13_+_24022963 | 0.70 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr1_+_25783801 | 0.70 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_10888762 | 0.70 |
ENSDART00000136049
|
syt10
|
synaptotagmin X |
| chr13_+_22119798 | 0.70 |
ENSDART00000173206
ENSDART00000078652 ENSDART00000165842 |
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr3_-_62380146 | 0.69 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr6_+_33537267 | 0.69 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr2_-_10188598 | 0.69 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr15_+_7054754 | 0.68 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr21_+_30194904 | 0.68 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr1_+_37195919 | 0.68 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr1_+_42225060 | 0.67 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr14_+_32022272 | 0.67 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.8 | 2.5 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.7 | 6.6 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.7 | 2.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 2.4 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.6 | 1.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.5 | 3.2 | GO:0006833 | water transport(GO:0006833) |
| 0.4 | 1.3 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.4 | 1.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 2.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.4 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.4 | 2.1 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.4 | 1.1 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 5.5 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.3 | 1.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.3 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 1.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 1.4 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 1.1 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 0.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.3 | 1.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 1.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.3 | 1.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.9 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.2 | 1.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 0.7 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.2 | 0.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.4 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.2 | 0.6 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 1.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 1.0 | GO:0051701 | interaction with host(GO:0051701) |
| 0.2 | 3.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.8 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.2 | 0.6 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 | 1.2 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.0 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 1.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 1.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 1.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.4 | GO:0045191 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.6 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.0 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.8 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 3.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 3.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.5 | GO:0048557 | embryonic digestive tract morphogenesis(GO:0048557) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.3 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 2.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.1 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 4.8 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 0.4 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.3 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.7 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.7 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 10.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.6 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 1.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.7 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:0060307 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 2.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.7 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.1 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) visceral serous pericardium development(GO:0061032) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:0070142 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.8 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.7 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.9 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.9 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.0 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.4 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 2.6 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 2.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.0 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.6 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.7 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 1.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0099590 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 2.8 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.0 | 0.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.0 | GO:0021610 | facial nerve morphogenesis(GO:0021610) facial nerve formation(GO:0021611) |
| 0.0 | 0.7 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.4 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.5 | 2.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 3.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 0.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 0.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 0.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.2 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.9 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 27.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 2.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 1.4 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.5 | 1.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 3.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 1.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.7 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.2 | 0.9 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.7 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.2 | 0.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 2.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 1.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 2.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 1.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.3 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.9 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.2 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 10.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 18.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 4.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.4 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 31.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0030552 | sodium channel activity(GO:0005272) cAMP binding(GO:0030552) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 7.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 2.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 6.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |