Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
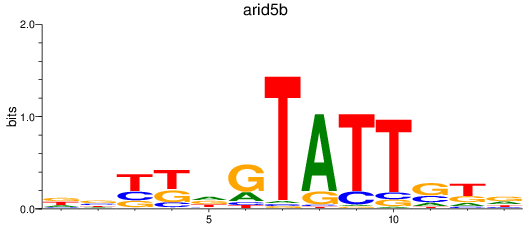
Results for arid5b
Z-value: 1.04
Transcription factors associated with arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid5b
|
ENSDARG00000037196 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid5b | dr11_v1_chr12_+_8373525_8373525 | -0.80 | 8.9e-03 | Click! |
Activity profile of arid5b motif
Sorted Z-values of arid5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_2322102 | 4.09 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr15_-_5624361 | 2.16 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr18_-_16953978 | 1.64 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr17_-_2578026 | 1.54 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2584423 | 1.54 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr25_+_33063762 | 1.52 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr25_+_9027831 | 1.52 |
ENSDART00000155855
|
im:7145024
|
im:7145024 |
| chr14_-_8940499 | 1.50 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr15_-_37589600 | 1.49 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr9_-_52814204 | 1.46 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr18_+_36782930 | 1.44 |
ENSDART00000004129
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr3_-_10634438 | 1.29 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr13_-_6252498 | 1.27 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr25_-_32363341 | 1.22 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr8_+_36500308 | 1.20 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr19_-_10330778 | 1.20 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr8_+_36500061 | 1.15 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr25_-_17918536 | 1.14 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr2_-_17114852 | 1.14 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr8_+_47342586 | 1.12 |
ENSDART00000007624
|
plch2a
|
phospholipase C, eta 2a |
| chr5_+_30596477 | 1.02 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr9_+_2020667 | 0.99 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr5_+_30596632 | 0.99 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr10_-_244745 | 0.99 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr20_+_33924235 | 0.97 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr18_+_27439680 | 0.94 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr12_+_19976400 | 0.93 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr21_+_3928947 | 0.92 |
ENSDART00000149777
|
setx
|
senataxin |
| chr3_-_40658820 | 0.90 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr16_+_53387085 | 0.89 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr23_-_24542156 | 0.87 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr23_-_27589754 | 0.86 |
ENSDART00000138381
ENSDART00000133721 |
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr17_+_10578823 | 0.83 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr16_-_42303856 | 0.82 |
ENSDART00000180030
|
ppox
|
protoporphyrinogen oxidase |
| chr5_+_33488860 | 0.82 |
ENSDART00000135571
|
si:dkey-238j22.1
|
si:dkey-238j22.1 |
| chr10_-_18463934 | 0.81 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr2_-_37744951 | 0.81 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr22_-_19102256 | 0.81 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr3_+_23047241 | 0.81 |
ENSDART00000103858
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr25_+_20694177 | 0.80 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr15_+_23356600 | 0.80 |
ENSDART00000113279
|
rnf26
|
ring finger protein 26 |
| chr2_+_42871831 | 0.79 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr20_+_54037138 | 0.79 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr5_-_69312533 | 0.76 |
ENSDART00000082614
ENSDART00000183098 |
smtnb
|
smoothelin b |
| chr9_-_14992730 | 0.74 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr19_-_32042105 | 0.74 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr5_-_72390259 | 0.74 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr23_+_20523617 | 0.74 |
ENSDART00000176404
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr2_+_31806602 | 0.74 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr11_+_42765963 | 0.73 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr2_+_42072231 | 0.71 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr7_+_13491452 | 0.68 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr5_-_69523816 | 0.68 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr15_-_1036878 | 0.67 |
ENSDART00000123844
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr5_+_64277604 | 0.66 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr15_-_1484795 | 0.65 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr14_-_21238046 | 0.64 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr17_+_28611746 | 0.62 |
ENSDART00000156711
ENSDART00000113300 |
mis18bp1
|
MIS18 binding protein 1 |
| chr1_+_58602506 | 0.62 |
ENSDART00000158425
|
si:ch73-221f6.1
|
si:ch73-221f6.1 |
| chr16_+_41293168 | 0.61 |
ENSDART00000191520
|
nek11
|
NIMA-related kinase 11 |
| chr15_-_1485086 | 0.61 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr9_-_2892045 | 0.59 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr23_+_3513201 | 0.59 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr8_+_22345282 | 0.59 |
ENSDART00000062254
|
MAD2L2
|
zgc:110299 |
| chr15_+_24572926 | 0.58 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr25_+_17244532 | 0.58 |
ENSDART00000050379
ENSDART00000171322 |
kdm7ab
|
lysine (K)-specific demethylase 7Ab |
| chr19_-_45650994 | 0.58 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr19_-_19379084 | 0.58 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr21_+_43172506 | 0.56 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr24_+_30392834 | 0.55 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr2_+_2967255 | 0.55 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr12_+_31783066 | 0.53 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr9_-_2892250 | 0.53 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr14_+_8940326 | 0.53 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr12_+_46841918 | 0.53 |
ENSDART00000157464
|
adkb
|
adenosine kinase b |
| chr23_+_44665230 | 0.51 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr2_-_37684641 | 0.51 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr23_-_29505645 | 0.50 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr23_-_27589508 | 0.49 |
ENSDART00000178404
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr2_+_42072689 | 0.49 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr9_+_14023386 | 0.48 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr10_+_2876020 | 0.47 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr2_-_50126337 | 0.45 |
ENSDART00000009347
|
exoc3
|
exocyst complex component 3 |
| chr11_-_18017918 | 0.45 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr10_+_2876378 | 0.42 |
ENSDART00000192083
|
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr3_-_60027255 | 0.42 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr2_+_54827391 | 0.42 |
ENSDART00000167269
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr5_-_29112956 | 0.41 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr15_-_34668485 | 0.40 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr11_+_42726712 | 0.39 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr4_+_20177526 | 0.38 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr6_-_6254432 | 0.38 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr17_-_7028418 | 0.35 |
ENSDART00000188305
ENSDART00000187895 |
sash1b
|
SAM and SH3 domain containing 1b |
| chr9_-_29985390 | 0.35 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr20_+_12702923 | 0.34 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr21_+_41839443 | 0.34 |
ENSDART00000010942
|
rnf14
|
ring finger protein 14 |
| chr11_+_7183025 | 0.34 |
ENSDART00000046670
ENSDART00000154009 ENSDART00000156974 ENSDART00000125619 |
thop1
|
thimet oligopeptidase 1 |
| chr10_+_35275965 | 0.33 |
ENSDART00000077404
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr23_-_27101600 | 0.33 |
ENSDART00000139231
|
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr12_+_18542954 | 0.32 |
ENSDART00000178845
ENSDART00000185149 ENSDART00000012743 |
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr16_-_27224000 | 0.31 |
ENSDART00000126347
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr11_-_36474306 | 0.31 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr6_-_9646275 | 0.31 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr13_-_30149973 | 0.30 |
ENSDART00000041515
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr15_+_21711671 | 0.30 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr19_+_13420884 | 0.29 |
ENSDART00000160209
|
si:ch211-204a13.2
|
si:ch211-204a13.2 |
| chr23_-_15878879 | 0.27 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr10_-_35410518 | 0.26 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr13_-_31166544 | 0.26 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr17_+_15213496 | 0.25 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr23_+_36730713 | 0.23 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr13_-_30150592 | 0.23 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr25_+_32157326 | 0.22 |
ENSDART00000112588
|
tjp1b
|
tight junction protein 1b |
| chr7_+_22792132 | 0.20 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr5_-_72178739 | 0.20 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr3_+_709393 | 0.18 |
ENSDART00000191970
|
AL929520.3
|
|
| chr22_-_22242884 | 0.18 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr24_-_7728409 | 0.18 |
ENSDART00000131392
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr5_-_1218801 | 0.18 |
ENSDART00000184398
|
CABZ01048143.1
|
|
| chr7_-_52709759 | 0.17 |
ENSDART00000136745
ENSDART00000131282 ENSDART00000073766 ENSDART00000161387 ENSDART00000174117 ENSDART00000174355 |
tcf12
|
transcription factor 12 |
| chr11_-_3954691 | 0.16 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr19_+_30884706 | 0.15 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr11_+_13224281 | 0.14 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr4_+_49196098 | 0.12 |
ENSDART00000150678
|
si:dkey-40n15.1
|
si:dkey-40n15.1 |
| chr13_+_22421151 | 0.12 |
ENSDART00000122687
|
opn4a
|
opsin 4a (melanopsin) |
| chr23_+_45734011 | 0.11 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr12_-_25097520 | 0.11 |
ENSDART00000158036
|
cript
|
cysteine-rich PDZ-binding protein |
| chr4_-_77377596 | 0.11 |
ENSDART00000186068
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr7_+_26998169 | 0.11 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr25_-_7925019 | 0.10 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr14_+_20351 | 0.10 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr2_-_3077027 | 0.10 |
ENSDART00000109319
|
arf1
|
ADP-ribosylation factor 1 |
| chr18_+_31410652 | 0.10 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr13_+_18523661 | 0.09 |
ENSDART00000034852
|
tlr4bb
|
toll-like receptor 4b, duplicate b |
| chr15_-_31390760 | 0.08 |
ENSDART00000190774
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr11_-_23322182 | 0.07 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr25_-_7925269 | 0.07 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr22_+_19311411 | 0.07 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr16_+_37876779 | 0.07 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr15_-_47812268 | 0.07 |
ENSDART00000190231
|
tomt
|
transmembrane O-methyltransferase |
| chr12_+_8074343 | 0.07 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr11_-_43002262 | 0.06 |
ENSDART00000172477
ENSDART00000181513 |
CABZ01069998.1
|
|
| chr18_-_7448047 | 0.05 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr16_-_14552199 | 0.05 |
ENSDART00000133368
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr10_-_28193642 | 0.04 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr10_+_40568735 | 0.04 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr7_+_29512673 | 0.04 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr17_-_50022827 | 0.03 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr11_+_1551603 | 0.03 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr25_-_2355107 | 0.02 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr16_+_27224068 | 0.00 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.3 | 1.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 1.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 3.1 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.5 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.6 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.6 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.9 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 1.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 1.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.1 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.8 | GO:0048264 | positive regulation of BMP signaling pathway(GO:0030513) determination of ventral identity(GO:0048264) |
| 0.0 | 0.4 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 2.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.0 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.2 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 1.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 3.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.4 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.6 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |