Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
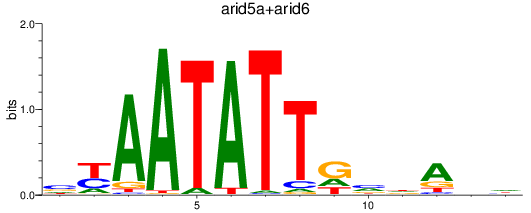
Results for arid5a+arid6
Z-value: 0.74
Transcription factors associated with arid5a+arid6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
|
arid5a
|
ENSDARG00000077120 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid5a | dr11_v1_chr8_+_52377516_52377516 | 0.25 | 5.2e-01 | Click! |
| arid6 | dr11_v1_chr21_+_11248448_11248448 | -0.05 | 8.9e-01 | Click! |
Activity profile of arid5a+arid6 motif
Sorted Z-values of arid5a+arid6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_39446247 | 0.45 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr19_+_23982466 | 0.43 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr9_-_11560110 | 0.42 |
ENSDART00000176941
|
cryba2b
|
crystallin, beta A2b |
| chr3_-_61205711 | 0.42 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr9_-_54001502 | 0.41 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr25_+_7494181 | 0.38 |
ENSDART00000165005
|
cat
|
catalase |
| chr14_+_51056605 | 0.36 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr11_-_24191928 | 0.35 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr16_-_9694822 | 0.35 |
ENSDART00000168748
|
COLEC10
|
si:ch211-93i7.4 |
| chr10_-_29903165 | 0.35 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr23_-_18609475 | 0.33 |
ENSDART00000104524
|
zgc:158296
|
zgc:158296 |
| chr15_+_44201056 | 0.31 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr25_+_459901 | 0.30 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr5_+_32221755 | 0.30 |
ENSDART00000125917
|
myhc4
|
myosin heavy chain 4 |
| chr22_-_24880824 | 0.30 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr25_+_33192796 | 0.30 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr16_+_28383758 | 0.29 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr9_-_42696408 | 0.29 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr24_-_17039638 | 0.29 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr24_-_7632187 | 0.28 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr1_+_46509176 | 0.28 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr24_-_36238054 | 0.28 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr18_+_25752592 | 0.28 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr12_+_16440708 | 0.28 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr6_-_42003780 | 0.27 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr22_+_25753972 | 0.26 |
ENSDART00000188417
|
AL929192.2
|
|
| chr6_-_18751696 | 0.26 |
ENSDART00000171537
|
tnrc6c2
|
trinucleotide repeat containing 6C2 |
| chr16_-_12173554 | 0.26 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr15_-_23647078 | 0.26 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr8_+_53344726 | 0.25 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr21_-_7940043 | 0.25 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr5_-_33886495 | 0.24 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr19_-_8940068 | 0.24 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr22_-_17611742 | 0.24 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr8_+_19977712 | 0.24 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr13_-_10431476 | 0.23 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr17_-_7218481 | 0.23 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr16_+_32729223 | 0.23 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr11_+_21910752 | 0.23 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr24_-_11446156 | 0.23 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr9_+_42606681 | 0.23 |
ENSDART00000191186
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr21_-_39639954 | 0.23 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr18_+_907266 | 0.22 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr14_+_35023923 | 0.22 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr3_-_12381271 | 0.22 |
ENSDART00000171068
ENSDART00000157475 |
coro7
|
coronin 7 |
| chr11_+_30647545 | 0.22 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr16_-_37372613 | 0.21 |
ENSDART00000124090
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr6_+_45932276 | 0.21 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr15_-_452347 | 0.21 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr16_-_17660594 | 0.21 |
ENSDART00000011936
|
ccdc106a
|
coiled-coil domain containing 106a |
| chr22_+_25704430 | 0.21 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr2_+_54696042 | 0.21 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr7_-_30925798 | 0.21 |
ENSDART00000149303
|
sord
|
sorbitol dehydrogenase |
| chr17_-_31058900 | 0.21 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr1_+_29068654 | 0.21 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr22_+_25693295 | 0.21 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr22_-_24858042 | 0.21 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr20_+_54024559 | 0.21 |
ENSDART00000130767
|
si:dkey-241l7.4
|
si:dkey-241l7.4 |
| chr6_+_51713076 | 0.21 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr14_+_35691889 | 0.20 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr19_+_41551543 | 0.20 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr1_+_10051763 | 0.20 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr8_+_36503797 | 0.20 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr12_-_22670279 | 0.20 |
ENSDART00000164888
|
zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr7_-_30926030 | 0.20 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr20_-_26066020 | 0.20 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr8_-_27858458 | 0.20 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr2_-_32769759 | 0.20 |
ENSDART00000178951
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr4_-_16353733 | 0.20 |
ENSDART00000186785
|
lum
|
lumican |
| chr2_-_32262287 | 0.20 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr13_-_18695289 | 0.20 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr25_+_20116407 | 0.20 |
ENSDART00000183615
ENSDART00000193243 |
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr10_+_26597990 | 0.19 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr22_-_18491813 | 0.19 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr21_+_36623162 | 0.19 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr22_+_25687525 | 0.19 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr1_-_338445 | 0.19 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr12_-_30558694 | 0.19 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr23_-_45407631 | 0.19 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr14_+_30398546 | 0.19 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr22_+_25681911 | 0.19 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr1_+_16073887 | 0.19 |
ENSDART00000160270
|
tusc3
|
tumor suppressor candidate 3 |
| chr25_-_29363934 | 0.18 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr15_-_26552393 | 0.18 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr2_+_37804235 | 0.17 |
ENSDART00000076321
|
ltb4r2b
|
leukotriene B4 receptor 2b |
| chr12_+_48220584 | 0.17 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr13_-_24448278 | 0.17 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr8_-_51365566 | 0.17 |
ENSDART00000182396
|
chmp7
|
charged multivesicular body protein 7 |
| chr17_-_10838434 | 0.17 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr12_-_47558276 | 0.17 |
ENSDART00000160260
|
grem2b
|
gremlin 2, DAN family BMP antagonist b |
| chr17_-_23895026 | 0.17 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr13_-_33022372 | 0.17 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr9_-_11549379 | 0.17 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr22_+_25715925 | 0.17 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr12_+_22588923 | 0.17 |
ENSDART00000184862
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr21_-_28340977 | 0.17 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr6_+_7322587 | 0.17 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr5_+_22177033 | 0.17 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr14_+_35013656 | 0.17 |
ENSDART00000164974
|
ebf3a
|
early B cell factor 3a |
| chr17_+_47116500 | 0.16 |
ENSDART00000186627
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr8_-_3963100 | 0.16 |
ENSDART00000139023
|
mtmr3
|
myotubularin related protein 3 |
| chr15_-_33172246 | 0.16 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr9_+_42546504 | 0.16 |
ENSDART00000192224
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr13_-_9895564 | 0.16 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr17_+_37227936 | 0.16 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr25_-_518656 | 0.16 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr6_+_39184236 | 0.16 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr18_+_31610744 | 0.16 |
ENSDART00000165660
|
dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr24_-_32582378 | 0.16 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr16_-_21047483 | 0.16 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr23_-_7826849 | 0.16 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr2_-_32768951 | 0.16 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr24_-_36910224 | 0.16 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr16_-_42390640 | 0.16 |
ENSDART00000193214
ENSDART00000102305 |
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr20_+_32552912 | 0.16 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr4_-_3353595 | 0.16 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr9_-_34191627 | 0.15 |
ENSDART00000142664
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr11_-_27501027 | 0.15 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr9_+_23900703 | 0.15 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr25_+_19530017 | 0.15 |
ENSDART00000144023
|
sh3gl3b
|
SH3-domain GRB2-like 3b |
| chr12_+_13091842 | 0.15 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr25_+_3099073 | 0.15 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr10_-_32851847 | 0.15 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr10_-_33156789 | 0.15 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr3_-_46410387 | 0.15 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr2_-_20715094 | 0.15 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr25_-_6049339 | 0.15 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr23_-_10048533 | 0.15 |
ENSDART00000166663
|
plxnb1a
|
plexin b1a |
| chr4_+_16794418 | 0.15 |
ENSDART00000079429
|
spx
|
spexin hormone |
| chr20_+_1960092 | 0.15 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr6_-_17999776 | 0.15 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr12_-_35988586 | 0.15 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr25_+_126174 | 0.14 |
ENSDART00000168913
|
cntn1a
|
contactin 1a |
| chr12_+_16233077 | 0.14 |
ENSDART00000152409
|
mpp3b
|
membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) |
| chr6_+_8315050 | 0.14 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr5_+_37837245 | 0.14 |
ENSDART00000171617
|
epd
|
ependymin |
| chr14_+_8127893 | 0.14 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr1_+_26667872 | 0.14 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr25_+_19105804 | 0.14 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr10_+_33393829 | 0.14 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr1_+_31864404 | 0.14 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr13_+_25455319 | 0.14 |
ENSDART00000145948
|
pkd2l1
|
polycystic kidney disease 2-like 1 |
| chr4_+_75200467 | 0.14 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr25_+_7423770 | 0.14 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr14_-_15990361 | 0.14 |
ENSDART00000168075
|
trim105
|
tripartite motif containing 105 |
| chr25_-_13381854 | 0.14 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr4_+_10754802 | 0.14 |
ENSDART00000181470
|
stab2
|
stabilin 2 |
| chr6_-_29051773 | 0.14 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr5_+_10084100 | 0.14 |
ENSDART00000109236
|
si:ch211-207k7.4
|
si:ch211-207k7.4 |
| chr21_+_974555 | 0.14 |
ENSDART00000130648
|
scarb2c
|
scavenger receptor class B, member 2c |
| chr19_-_47455944 | 0.14 |
ENSDART00000190005
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr15_-_31067589 | 0.14 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr9_+_42607138 | 0.13 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr20_-_14114078 | 0.13 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr11_-_38083397 | 0.13 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr7_+_71664624 | 0.13 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr10_+_21780250 | 0.13 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr1_-_18592068 | 0.13 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr3_+_31039923 | 0.13 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr17_-_38291065 | 0.13 |
ENSDART00000152056
|
pax9
|
paired box 9 |
| chr25_+_4837915 | 0.13 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr2_+_36109002 | 0.13 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr1_-_22412042 | 0.13 |
ENSDART00000074678
|
chrnb3a
|
cholinergic receptor, nicotinic, beta polypeptide 3a |
| chr13_-_22907260 | 0.13 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr7_+_39399747 | 0.13 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr13_-_12389748 | 0.13 |
ENSDART00000141606
|
commd8
|
COMM domain containing 8 |
| chr19_+_32257472 | 0.13 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr16_-_52821023 | 0.13 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr22_+_2959879 | 0.13 |
ENSDART00000185354
ENSDART00000063545 |
impact
|
impact RWD domain protein |
| chr11_-_42472941 | 0.13 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr2_+_1999255 | 0.13 |
ENSDART00000153597
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr21_-_40173821 | 0.13 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr13_+_15354012 | 0.13 |
ENSDART00000121453
|
loxl3b
|
lysyl oxidase-like 3b |
| chr7_+_21275152 | 0.12 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr14_-_29905962 | 0.12 |
ENSDART00000142605
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr11_+_30244356 | 0.12 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr15_+_32387063 | 0.12 |
ENSDART00000154210
ENSDART00000156525 |
si:ch211-162k9.5
|
si:ch211-162k9.5 |
| chr24_+_7631797 | 0.12 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr12_+_23850661 | 0.12 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr9_+_4306122 | 0.12 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr15_-_42039192 | 0.12 |
ENSDART00000004338
|
epha4l
|
eph receptor A4, like |
| chr17_-_29312506 | 0.12 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr21_+_22124736 | 0.12 |
ENSDART00000130179
ENSDART00000172573 |
cul5b
|
cullin 5b |
| chr12_+_36428052 | 0.12 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr18_+_40471826 | 0.12 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr16_+_10972733 | 0.12 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr15_-_12360409 | 0.12 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr11_+_36408636 | 0.12 |
ENSDART00000182098
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr22_+_737211 | 0.12 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr2_-_24996441 | 0.12 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr12_-_19102736 | 0.12 |
ENSDART00000180364
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr5_-_67895656 | 0.12 |
ENSDART00000158917
|
abhd10a
|
abhydrolase domain containing 10a |
| chr25_-_35664817 | 0.12 |
ENSDART00000148718
|
lrrk2
|
leucine-rich repeat kinase 2 |
| chr3_-_13461361 | 0.12 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr2_+_40294313 | 0.12 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr12_+_2665081 | 0.12 |
ENSDART00000147532
|
rbp3
|
retinol binding protein 3 |
| chr24_+_23791758 | 0.12 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr7_-_8504355 | 0.11 |
ENSDART00000173067
|
loc564660
|
hypothetical protein LOC564660 |
| chr7_-_30177691 | 0.11 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr13_-_28265919 | 0.11 |
ENSDART00000057565
|
sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5a+arid6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.3 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.3 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:2000320 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.3 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.5 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.0 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.4 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0098754 | detoxification(GO:0098754) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |