Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
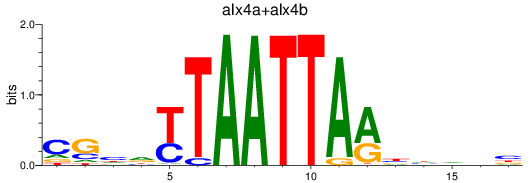
Results for alx4a+alx4b
Z-value: 0.62
Transcription factors associated with alx4a+alx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx4b
|
ENSDARG00000074442 | ALX homeobox 4b |
|
alx4a
|
ENSDARG00000088332 | ALX homeobox 4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx4b | dr11_v1_chr18_+_38321039_38321039 | 0.84 | 4.5e-03 | Click! |
| alx4a | dr11_v1_chr7_-_26924903_26924903 | 0.77 | 1.5e-02 | Click! |
Activity profile of alx4a+alx4b motif
Sorted Z-values of alx4a+alx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 2.00 |
ENSDART00000101923
|
CR391987.1
|
|
| chr17_+_15433671 | 1.35 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433518 | 1.35 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr3_-_32818607 | 1.23 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr25_+_29161609 | 1.19 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr4_+_9669717 | 1.19 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr12_-_35830625 | 1.17 |
ENSDART00000180028
|
CU459056.1
|
|
| chr25_+_31276842 | 1.15 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr20_+_19512727 | 1.10 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr3_-_55128258 | 1.06 |
ENSDART00000101734
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr9_-_22129788 | 0.98 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr4_+_16885854 | 0.88 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr14_-_413273 | 0.88 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr3_+_28939759 | 0.87 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr8_+_31435452 | 0.85 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr21_+_25236297 | 0.80 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr13_-_29420885 | 0.79 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr14_-_4145594 | 0.78 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr5_-_41494831 | 0.72 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr9_-_14504834 | 0.72 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr21_+_28958471 | 0.71 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_+_10855158 | 0.70 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr24_-_40860603 | 0.70 |
ENSDART00000188032
|
CU633479.7
|
|
| chr23_-_23401305 | 0.69 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr17_-_12336987 | 0.66 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr18_-_48983690 | 0.65 |
ENSDART00000182359
|
FO681288.3
|
|
| chr12_+_13118540 | 0.64 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr18_+_2837563 | 0.64 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr11_+_30057762 | 0.64 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr21_+_45841731 | 0.63 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr6_-_607063 | 0.62 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr22_+_11775269 | 0.62 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_+_12915498 | 0.62 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr6_-_7720332 | 0.61 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr19_+_43297546 | 0.61 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr2_-_38000276 | 0.61 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr22_+_16535575 | 0.61 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr16_-_28856112 | 0.60 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr4_-_9891874 | 0.59 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr15_-_22074315 | 0.59 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr15_-_12011390 | 0.59 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr14_+_45406299 | 0.58 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr15_-_5815006 | 0.58 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr7_-_73752955 | 0.58 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr7_-_66877058 | 0.57 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr3_-_53533128 | 0.56 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr17_+_41992054 | 0.56 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr6_+_35362225 | 0.56 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr8_+_7359294 | 0.55 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr24_-_7697274 | 0.54 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr23_-_42232124 | 0.54 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr21_-_19314618 | 0.54 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr11_-_7261717 | 0.53 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr15_+_23799461 | 0.53 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr8_+_34731982 | 0.53 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr3_+_28953274 | 0.52 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr10_-_14556978 | 0.52 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr14_-_49063157 | 0.51 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr24_+_21621654 | 0.51 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr2_+_35728033 | 0.51 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr8_+_23738122 | 0.50 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr23_+_44741500 | 0.49 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr10_-_43771447 | 0.49 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr3_-_12930217 | 0.48 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr13_-_22862133 | 0.48 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr6_+_47424266 | 0.48 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr23_-_21446985 | 0.47 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr15_-_3736773 | 0.47 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr18_+_21408794 | 0.47 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr16_+_6864608 | 0.47 |
ENSDART00000078306
|
arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr25_-_13569234 | 0.47 |
ENSDART00000189645
|
fa2h
|
fatty acid 2-hydroxylase |
| chr24_-_37955993 | 0.46 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr1_-_669717 | 0.46 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr2_-_30668580 | 0.46 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr18_-_48547564 | 0.45 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr15_-_47848544 | 0.44 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr10_-_26744131 | 0.44 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr24_-_25691020 | 0.44 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr1_+_127250 | 0.43 |
ENSDART00000003463
|
f7i
|
coagulation factor VIIi |
| chr20_-_23439011 | 0.43 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr11_-_45141309 | 0.43 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr1_-_46981134 | 0.43 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr1_+_59293873 | 0.42 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr5_+_71802014 | 0.42 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr20_-_9436521 | 0.42 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr22_-_23748284 | 0.41 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr16_-_12173554 | 0.41 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr5_+_64739762 | 0.41 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr3_-_57666518 | 0.41 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr6_-_58764672 | 0.40 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr25_+_5039050 | 0.40 |
ENSDART00000154700
|
parvb
|
parvin, beta |
| chr8_+_6863462 | 0.40 |
ENSDART00000064163
|
nefmb
|
neurofilament, medium polypeptide b |
| chr4_-_4834347 | 0.40 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr11_+_34760628 | 0.40 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr4_+_43522680 | 0.40 |
ENSDART00000182252
|
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr2_-_56348727 | 0.40 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr11_-_1509773 | 0.40 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr18_+_48423973 | 0.39 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr5_-_54672763 | 0.39 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr10_-_42882215 | 0.39 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr9_-_53666031 | 0.39 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr17_+_51627209 | 0.39 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr7_-_33960170 | 0.39 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr17_-_29224908 | 0.39 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr14_+_24284756 | 0.38 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr7_+_59020972 | 0.38 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr3_-_60175470 | 0.38 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr6_+_2097690 | 0.37 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr19_+_43359075 | 0.36 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr13_+_255067 | 0.36 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr12_-_9294819 | 0.36 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr22_-_10121880 | 0.36 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr7_-_30174882 | 0.35 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr6_-_11768198 | 0.35 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr14_+_5385855 | 0.35 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr20_-_45812144 | 0.35 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr16_-_43344859 | 0.35 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr25_+_11008419 | 0.35 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr7_+_6652967 | 0.35 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr6_+_25257728 | 0.35 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr11_-_45138857 | 0.34 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr5_+_46481586 | 0.34 |
ENSDART00000144973
|
vcana
|
versican a |
| chr23_-_3681026 | 0.34 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr25_-_35360096 | 0.34 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr14_-_7137808 | 0.34 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr3_-_19368435 | 0.34 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr5_-_20205075 | 0.34 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr22_-_12160283 | 0.34 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr11_+_26476153 | 0.34 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr6_+_52350443 | 0.33 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr2_+_2223837 | 0.33 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr24_-_38657683 | 0.33 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr1_+_6172786 | 0.33 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr4_+_22480169 | 0.33 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr12_+_31713239 | 0.33 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr4_+_21129752 | 0.33 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr15_-_21014270 | 0.33 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr3_+_25154078 | 0.32 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr17_+_38476300 | 0.32 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr23_+_43255328 | 0.32 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr22_-_10482253 | 0.32 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr18_-_46258612 | 0.32 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr5_+_11840905 | 0.32 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr1_-_25966068 | 0.32 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr8_+_23521974 | 0.31 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr16_-_11798994 | 0.31 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr3_+_33341640 | 0.31 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr6_-_46941245 | 0.31 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr5_+_36752943 | 0.31 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr1_-_43915423 | 0.30 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr14_-_858985 | 0.30 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr2_+_11685742 | 0.30 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr8_+_28259347 | 0.30 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr19_-_5699703 | 0.30 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr2_-_27329667 | 0.30 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr19_+_43780970 | 0.30 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr15_-_47865063 | 0.30 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr4_+_57881965 | 0.30 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr21_+_22840246 | 0.30 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr12_-_31794908 | 0.30 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr6_-_39270851 | 0.30 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr11_+_41242644 | 0.30 |
ENSDART00000172008
|
pax7a
|
paired box 7a |
| chr16_-_12173399 | 0.29 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr15_+_45640906 | 0.29 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr23_+_26733232 | 0.29 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr4_+_16715267 | 0.29 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr15_+_9327252 | 0.29 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr8_+_24745041 | 0.29 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr17_-_36896560 | 0.29 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr4_-_12907826 | 0.29 |
ENSDART00000182923
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr20_-_9760424 | 0.29 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_+_20471315 | 0.28 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr19_-_26869103 | 0.28 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr23_-_20051369 | 0.28 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr18_+_24919614 | 0.28 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr19_+_156757 | 0.28 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr2_+_20406399 | 0.28 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr10_-_35236949 | 0.28 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr8_+_49778486 | 0.28 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr7_+_39634873 | 0.28 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr19_-_10810006 | 0.28 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr9_-_1703761 | 0.28 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr5_+_40485503 | 0.28 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr20_+_53441935 | 0.28 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr5_-_12219572 | 0.27 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr19_+_12237945 | 0.27 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr4_-_9909371 | 0.27 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr7_-_71829649 | 0.27 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr4_-_1908179 | 0.27 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr8_-_4596662 | 0.27 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr4_-_76488854 | 0.27 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr1_+_45663727 | 0.26 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr1_+_52560549 | 0.26 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr6_-_54815886 | 0.26 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr18_-_34549721 | 0.26 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr8_-_12867434 | 0.26 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr22_+_7738966 | 0.26 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr8_-_30979494 | 0.26 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr11_-_2131280 | 0.26 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr4_+_72723304 | 0.25 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx4a+alx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.3 | 0.8 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.2 | 0.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.6 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 0.7 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.2 | 0.5 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.3 | GO:0070169 | positive regulation of bone mineralization(GO:0030501) positive regulation of ossification(GO:0045778) positive regulation of biomineral tissue development(GO:0070169) |
| 0.1 | 0.4 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.3 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.3 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.2 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.2 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.5 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.5 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.3 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.2 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.4 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 2.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.4 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.2 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.1 | GO:0032602 | chemokine production(GO:0032602) negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.0 | 0.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.6 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.0 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.3 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 2.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.5 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.4 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0070915 | bioactive lipid receptor activity(GO:0045125) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0022821 | calcium, potassium:sodium antiporter activity(GO:0008273) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |