Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
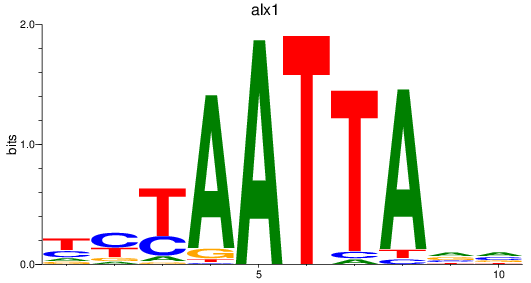
Results for alx1
Z-value: 0.86
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000110530 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000115230 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr11_v1_chr18_+_16246806_16246806 | -0.11 | 7.9e-01 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 1.25 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr2_+_50608099 | 0.92 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_-_5103141 | 0.75 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_-_45060241 | 0.70 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr7_-_51773166 | 0.66 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr1_-_43905252 | 0.64 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr2_+_37227011 | 0.64 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr8_+_45334255 | 0.61 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr12_-_14143344 | 0.60 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr16_+_39159752 | 0.54 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr19_-_5103313 | 0.53 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_+_44804685 | 0.51 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr4_-_13580348 | 0.49 |
ENSDART00000067160
|
opn1sw1
|
opsin 1 (cone pigments), short-wave-sensitive 1 |
| chr21_-_14251306 | 0.48 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr9_+_29548195 | 0.47 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr20_-_9436521 | 0.47 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr20_+_32523576 | 0.46 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr15_+_47903864 | 0.44 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr17_-_41798856 | 0.44 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr22_-_17653143 | 0.43 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr23_+_20563779 | 0.43 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr22_+_17828267 | 0.42 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr15_-_34408777 | 0.42 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr22_+_34784075 | 0.41 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr21_-_14174786 | 0.40 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr23_-_27571667 | 0.40 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr20_-_45062514 | 0.40 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr21_+_25777425 | 0.39 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_25217347 | 0.38 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_-_32157865 | 0.38 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr21_-_32060993 | 0.38 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr21_+_3093419 | 0.37 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr14_+_44860335 | 0.37 |
ENSDART00000091620
ENSDART00000173043 ENSDART00000091625 |
atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr16_+_47207691 | 0.36 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr13_+_38430466 | 0.36 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr5_+_57924611 | 0.36 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr15_-_6247775 | 0.36 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr5_-_20491198 | 0.35 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr11_-_40418975 | 0.35 |
ENSDART00000086296
|
trim62
|
tripartite motif containing 62 |
| chr25_+_7504314 | 0.35 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr24_-_7697274 | 0.34 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr10_+_16911951 | 0.34 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr4_-_14315855 | 0.34 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr14_-_8940499 | 0.33 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr9_+_50001746 | 0.33 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr15_-_33172246 | 0.33 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr11_+_35171406 | 0.33 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr19_-_5865766 | 0.33 |
ENSDART00000191007
|
LO018585.1
|
|
| chr18_+_1703984 | 0.32 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr4_+_21129752 | 0.32 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr16_+_27349585 | 0.32 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr19_+_10339538 | 0.31 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr21_-_36453594 | 0.31 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr7_+_25126629 | 0.31 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr17_+_26722904 | 0.31 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr17_-_40956035 | 0.31 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr24_+_1023839 | 0.30 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr23_+_28092083 | 0.29 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr1_+_18811679 | 0.29 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr10_-_21362071 | 0.29 |
ENSDART00000125167
|
avd
|
avidin |
| chr15_-_44601331 | 0.29 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr25_+_33849647 | 0.29 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr10_-_11385155 | 0.29 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr19_-_5805923 | 0.29 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr6_-_50203682 | 0.29 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr2_-_42871286 | 0.29 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr5_-_13206878 | 0.28 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr11_+_12052791 | 0.28 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr5_-_28915130 | 0.28 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr21_-_25722834 | 0.28 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr16_-_28658341 | 0.28 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr9_+_12887491 | 0.27 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr11_+_31864921 | 0.27 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr16_-_34401412 | 0.27 |
ENSDART00000054020
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr25_-_27564205 | 0.27 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr22_-_17652914 | 0.27 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr22_+_4488454 | 0.27 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr3_+_32553714 | 0.27 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr9_-_14683574 | 0.27 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr7_-_71389375 | 0.26 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr21_+_34088377 | 0.26 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr8_+_11425048 | 0.26 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr12_-_13549538 | 0.26 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr6_+_29693492 | 0.26 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr24_+_12835935 | 0.26 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr6_+_9870192 | 0.26 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr24_+_16547035 | 0.26 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr21_-_18993110 | 0.26 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr18_-_16801033 | 0.26 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr13_+_38817871 | 0.26 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr17_-_200316 | 0.26 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr24_-_31452875 | 0.25 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr6_-_46861676 | 0.25 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr20_-_40755614 | 0.25 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr5_+_27137473 | 0.25 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr23_+_40460333 | 0.25 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr18_-_43884044 | 0.25 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_+_29997043 | 0.25 |
ENSDART00000139566
ENSDART00000151848 |
rbm33b
|
RNA binding motif protein 33b |
| chr15_+_31344472 | 0.24 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr7_+_23292133 | 0.24 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr5_+_51594209 | 0.24 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr10_-_32494499 | 0.24 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr6_-_30839763 | 0.24 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr9_+_45428041 | 0.24 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr18_-_399554 | 0.24 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr23_+_24272421 | 0.24 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr9_+_29548630 | 0.24 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr12_-_30358836 | 0.24 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr22_+_39096911 | 0.24 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr24_+_17068724 | 0.24 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr7_+_22313533 | 0.24 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr18_+_15644559 | 0.23 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr12_-_6818676 | 0.23 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr21_-_39177564 | 0.23 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr21_-_16632808 | 0.23 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr9_+_34641237 | 0.23 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr16_-_46579936 | 0.23 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr25_-_13381854 | 0.23 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr2_-_3403020 | 0.23 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr10_-_32494304 | 0.23 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr2_-_15324837 | 0.23 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_+_518546 | 0.23 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr24_-_5713799 | 0.23 |
ENSDART00000137293
|
dia1b
|
deleted in autism 1b |
| chr25_+_5604512 | 0.23 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr14_+_30773764 | 0.22 |
ENSDART00000186961
|
atl3
|
atlastin 3 |
| chr13_-_32726178 | 0.22 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr16_+_2820340 | 0.22 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr9_-_50001606 | 0.22 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr25_+_3035384 | 0.22 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr1_+_18920213 | 0.22 |
ENSDART00000193777
|
rnf38
|
ring finger protein 38 |
| chr6_-_12172424 | 0.22 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr15_+_44366556 | 0.22 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr19_-_30510259 | 0.22 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr7_-_54217547 | 0.22 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr1_+_21937201 | 0.22 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr2_+_42871831 | 0.22 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr10_-_27049170 | 0.22 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr21_-_28640316 | 0.21 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr5_+_61301525 | 0.21 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr7_+_26649319 | 0.21 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr13_+_23095228 | 0.21 |
ENSDART00000189068
ENSDART00000188624 |
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr24_+_5208171 | 0.21 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr7_+_48761875 | 0.21 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr12_+_45677293 | 0.21 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr5_+_29794058 | 0.21 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr24_+_5912635 | 0.21 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr16_-_16120941 | 0.21 |
ENSDART00000131227
|
ankib1b
|
ankyrin repeat and IBR domain containing 1b |
| chr5_-_61797220 | 0.21 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr2_+_20332044 | 0.21 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr22_-_10486477 | 0.21 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr2_+_38002717 | 0.20 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr8_-_21142550 | 0.20 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr12_+_22407852 | 0.20 |
ENSDART00000178840
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr3_-_25268751 | 0.20 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr4_+_11723852 | 0.20 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_-_25034411 | 0.20 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr12_-_30359498 | 0.20 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr22_+_20427170 | 0.20 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr12_-_6880694 | 0.20 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr19_-_32710922 | 0.20 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr22_-_24818066 | 0.20 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr10_-_21362320 | 0.19 |
ENSDART00000189789
|
avd
|
avidin |
| chr11_-_1550709 | 0.19 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr12_-_30359031 | 0.19 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr19_+_41464870 | 0.19 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr22_-_14739491 | 0.19 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr13_+_40501455 | 0.19 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr23_-_36823932 | 0.19 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr21_-_36453417 | 0.19 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr23_+_45584223 | 0.19 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr2_+_6963296 | 0.19 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr4_-_12766418 | 0.19 |
ENSDART00000024312
|
dera
|
deoxyribose-phosphate aldolase (putative) |
| chr23_-_30954738 | 0.19 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr15_+_22867174 | 0.19 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr10_-_31015535 | 0.19 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr5_+_31283576 | 0.19 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr21_+_34088110 | 0.19 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr1_+_35985813 | 0.19 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr8_+_25145464 | 0.19 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr5_+_35786141 | 0.19 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr8_+_3820134 | 0.18 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr12_-_33579873 | 0.18 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr15_-_9272328 | 0.18 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr22_-_20924564 | 0.18 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr14_-_2933185 | 0.18 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr3_+_25154078 | 0.18 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr2_+_25198648 | 0.18 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_+_6010570 | 0.18 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr5_+_58421536 | 0.18 |
ENSDART00000173410
|
ccdc15
|
coiled-coil domain containing 15 |
| chr6_-_12900154 | 0.18 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr20_+_16639848 | 0.18 |
ENSDART00000063944
ENSDART00000152359 |
tmem30ab
|
transmembrane protein 30Ab |
| chr23_-_32162810 | 0.18 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_-_977849 | 0.17 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr25_+_31227747 | 0.17 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr14_+_35428152 | 0.17 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr13_-_31017960 | 0.17 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr8_+_41037541 | 0.17 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr21_-_22737228 | 0.17 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr25_+_31277415 | 0.17 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr18_-_1185772 | 0.17 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.7 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.9 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.9 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.4 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.2 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.0 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.0 | 0.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.1 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.7 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |