Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
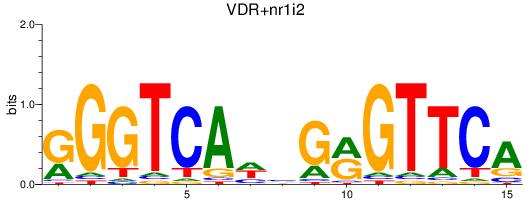
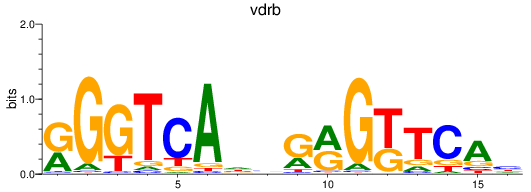
Results for VDR+nr1i2_vdrb
Z-value: 1.08
Transcription factors associated with VDR+nr1i2_vdrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1i2
|
ENSDARG00000029766 | nuclear receptor subfamily 1, group I, member 2 |
|
VDR
|
ENSDARG00000112923 | vitamin D receptor |
|
vdrb
|
ENSDARG00000070721 | vitamin D receptor b |
|
vdrb
|
ENSDARG00000114864 | vitamin D receptor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vdrb | dr11_v1_chr6_-_39005133_39005133 | 0.23 | 5.5e-01 | Click! |
| nr1i2 | dr11_v1_chr9_+_9502610_9502610 | 0.23 | 5.5e-01 | Click! |
| CABZ01088036.1 | dr11_v1_chr23_+_45785563_45785563 | 0.18 | 6.3e-01 | Click! |
Activity profile of VDR+nr1i2_vdrb motif
Sorted Z-values of VDR+nr1i2_vdrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_20148469 | 2.83 |
ENSDART00000134476
|
si:ch211-155k24.1
|
si:ch211-155k24.1 |
| chr3_-_34656745 | 1.72 |
ENSDART00000144824
|
znfl1
|
zinc finger-like gene 1 |
| chr9_+_15837398 | 1.68 |
ENSDART00000141063
|
si:dkey-103d23.5
|
si:dkey-103d23.5 |
| chr8_+_23245596 | 1.67 |
ENSDART00000143313
|
si:ch211-196c10.11
|
si:ch211-196c10.11 |
| chr20_+_34374735 | 1.66 |
ENSDART00000144090
|
si:dkeyp-11g8.3
|
si:dkeyp-11g8.3 |
| chr9_-_31763246 | 1.62 |
ENSDART00000114742
|
si:dkey-250i3.3
|
si:dkey-250i3.3 |
| chr13_+_45332203 | 1.42 |
ENSDART00000110466
|
si:ch211-168h21.3
|
si:ch211-168h21.3 |
| chr19_-_9776075 | 0.87 |
ENSDART00000140332
|
si:dkey-14o18.1
|
si:dkey-14o18.1 |
| chr6_+_50451337 | 0.84 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr5_+_22459087 | 0.63 |
ENSDART00000134781
|
BX546499.1
|
|
| chr3_-_29977495 | 0.54 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr23_-_44723102 | 0.53 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr25_+_17871089 | 0.51 |
ENSDART00000133725
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr13_-_36418921 | 0.51 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr7_-_24995631 | 0.50 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr21_+_22878991 | 0.50 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr14_+_35383267 | 0.43 |
ENSDART00000143818
|
clint1a
|
clathrin interactor 1a |
| chr21_+_22878834 | 0.42 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr14_+_22076596 | 0.42 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr25_+_17870799 | 0.42 |
ENSDART00000136797
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr21_+_29179887 | 0.41 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr17_-_26868169 | 0.39 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr7_-_24236364 | 0.39 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr9_+_21306902 | 0.38 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr20_+_17581881 | 0.38 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr10_+_15603082 | 0.36 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr21_+_5589923 | 0.34 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr9_-_28616436 | 0.32 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr18_+_18879733 | 0.31 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr6_+_59840360 | 0.31 |
ENSDART00000154647
|
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr21_-_19918286 | 0.30 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_-_24101897 | 0.29 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr22_+_35205968 | 0.29 |
ENSDART00000150467
|
tsc22d2
|
TSC22 domain family 2 |
| chr7_+_47243564 | 0.28 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr5_-_54712159 | 0.28 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr23_+_16430559 | 0.27 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr1_+_54626491 | 0.26 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr10_+_7564106 | 0.24 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr5_+_41476443 | 0.23 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_+_42691210 | 0.23 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr15_-_25198107 | 0.23 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr3_+_19336286 | 0.23 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr7_+_24069421 | 0.22 |
ENSDART00000034218
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr16_+_40954481 | 0.22 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr7_+_12931396 | 0.22 |
ENSDART00000184664
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr10_+_15024772 | 0.22 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr10_+_25369859 | 0.22 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr23_+_42454292 | 0.22 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| chr5_+_29160132 | 0.22 |
ENSDART00000088827
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr10_+_15025006 | 0.22 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr12_+_20506197 | 0.21 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr15_+_2421729 | 0.21 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr5_-_42544522 | 0.21 |
ENSDART00000157968
|
BX510949.1
|
|
| chr8_+_50742975 | 0.21 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr6_-_13498745 | 0.20 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr2_+_33051730 | 0.20 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr1_-_23595779 | 0.20 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr3_-_32912787 | 0.19 |
ENSDART00000156149
|
g6pcb
|
glucose-6-phosphatase b, catalytic subunit |
| chr14_+_31473866 | 0.19 |
ENSDART00000173088
|
ccdc160
|
coiled-coil domain containing 160 |
| chr15_-_24883956 | 0.19 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr14_-_41468892 | 0.19 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr22_-_4878012 | 0.18 |
ENSDART00000110947
ENSDART00000147431 |
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr5_+_23624684 | 0.18 |
ENSDART00000051539
|
cx27.5
|
connexin 27.5 |
| chr2_+_33052169 | 0.18 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr22_+_26798853 | 0.18 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr12_-_25217217 | 0.17 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr4_-_20511595 | 0.17 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr4_-_20177868 | 0.17 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr15_+_1653779 | 0.17 |
ENSDART00000021299
|
nmd3
|
NMD3 ribosome export adaptor |
| chr14_-_31856819 | 0.16 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_30528239 | 0.16 |
ENSDART00000169290
|
znf1052
|
zinc finger protein 1052 |
| chr14_-_33872092 | 0.16 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr12_+_18663154 | 0.16 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr15_+_20530649 | 0.16 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr21_+_38089036 | 0.16 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr5_+_70271799 | 0.15 |
ENSDART00000101316
|
znf618
|
zinc finger protein 618 |
| chr19_+_29798064 | 0.15 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr4_-_29833326 | 0.15 |
ENSDART00000150596
|
si:ch211-231i17.4
|
si:ch211-231i17.4 |
| chr5_+_51079504 | 0.15 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr20_+_49081967 | 0.15 |
ENSDART00000112689
|
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr12_+_24060894 | 0.15 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr4_-_43388943 | 0.15 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr20_+_33744065 | 0.15 |
ENSDART00000015000
|
henmt1
|
HEN methyltransferase 1 |
| chr10_-_17550931 | 0.15 |
ENSDART00000145077
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr22_+_34694217 | 0.14 |
ENSDART00000111793
|
hykk.1
|
hydroxylysine kinase, tandem duplicate 1 |
| chr19_-_31576321 | 0.14 |
ENSDART00000103612
|
tdp2b
|
tyrosyl-DNA phosphodiesterase 2b |
| chr13_+_18507592 | 0.14 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr22_+_24389135 | 0.14 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr2_+_10134345 | 0.14 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr14_+_8715527 | 0.14 |
ENSDART00000170754
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr12_-_13556536 | 0.14 |
ENSDART00000166396
|
CR354395.1
|
|
| chr15_-_8665662 | 0.14 |
ENSDART00000090675
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr5_-_1076431 | 0.13 |
ENSDART00000168336
|
si:zfos-128g4.1
|
si:zfos-128g4.1 |
| chr12_-_30359031 | 0.13 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr13_+_22731356 | 0.13 |
ENSDART00000133064
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr15_-_28805493 | 0.13 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr18_+_20482369 | 0.13 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr8_+_8671229 | 0.13 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr3_+_36366053 | 0.13 |
ENSDART00000181140
|
rgs9a
|
regulator of G protein signaling 9a |
| chr23_-_18595020 | 0.13 |
ENSDART00000187032
ENSDART00000191047 |
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr3_-_24681404 | 0.12 |
ENSDART00000161612
|
BX569789.1
|
|
| chr16_-_21692024 | 0.12 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr11_-_36263886 | 0.12 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr20_+_48100261 | 0.12 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr24_-_21498802 | 0.12 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr1_+_54901028 | 0.12 |
ENSDART00000137352
|
zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr5_+_37729207 | 0.11 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr2_+_16710889 | 0.11 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr20_+_20731633 | 0.11 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr24_+_7742441 | 0.11 |
ENSDART00000189956
|
zgc:56095
|
zgc:56095 |
| chr15_+_24756860 | 0.11 |
ENSDART00000156424
ENSDART00000078035 |
cpda
|
carboxypeptidase D, a |
| chr6_-_43677125 | 0.10 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr19_-_35400819 | 0.10 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr11_+_2642013 | 0.10 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr21_-_15200556 | 0.10 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr6_-_39080630 | 0.10 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr10_+_7563755 | 0.10 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr6_-_25369295 | 0.10 |
ENSDART00000191475
|
PKN2 (1 of many)
|
zgc:153916 |
| chr16_-_22781446 | 0.10 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr21_+_25216397 | 0.10 |
ENSDART00000130970
|
sycn.3
|
syncollin, tandem duplicate 3 |
| chr24_+_38155830 | 0.09 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr21_-_1012269 | 0.09 |
ENSDART00000159835
|
CABZ01057159.1
|
|
| chr17_-_50071748 | 0.09 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr6_-_11812224 | 0.09 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr10_-_373575 | 0.09 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr1_+_39865748 | 0.09 |
ENSDART00000131954
ENSDART00000130270 |
irf2a
|
interferon regulatory factor 2a |
| chr20_+_29209926 | 0.09 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_+_12744083 | 0.09 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr11_-_3907937 | 0.09 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr22_-_4880603 | 0.08 |
ENSDART00000134055
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr8_+_16990120 | 0.08 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr4_-_75172216 | 0.08 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr16_-_30878521 | 0.08 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr10_+_428269 | 0.08 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr7_+_25000060 | 0.08 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr16_+_42772678 | 0.08 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr23_-_38054 | 0.08 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr14_-_32486757 | 0.08 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr8_+_21133307 | 0.08 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr2_-_24898226 | 0.08 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr6_-_9630081 | 0.07 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr20_+_23960525 | 0.07 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr21_+_39197418 | 0.07 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr6_-_9922266 | 0.07 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr7_-_12065668 | 0.07 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr3_+_62356578 | 0.07 |
ENSDART00000157030
|
iqck
|
IQ motif containing K |
| chr12_-_30358836 | 0.07 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr5_-_69716501 | 0.07 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr1_+_54687616 | 0.07 |
ENSDART00000146677
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr12_+_48581588 | 0.07 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr24_+_19986432 | 0.07 |
ENSDART00000184402
|
CR381686.5
|
|
| chr14_+_30753666 | 0.07 |
ENSDART00000009385
ENSDART00000137782 ENSDART00000172778 |
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr9_+_18576047 | 0.06 |
ENSDART00000145174
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr1_-_56951369 | 0.06 |
ENSDART00000152294
|
si:ch211-1f22.2
|
si:ch211-1f22.2 |
| chr20_-_24165509 | 0.06 |
ENSDART00000124919
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr2_-_22575851 | 0.06 |
ENSDART00000182932
|
FQ377628.2
|
|
| chr4_-_13921185 | 0.06 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr16_+_21790870 | 0.06 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr3_+_36366577 | 0.06 |
ENSDART00000151203
|
rgs9a
|
regulator of G protein signaling 9a |
| chr24_+_17345521 | 0.06 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr14_+_22297257 | 0.06 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr4_-_18454591 | 0.06 |
ENSDART00000185200
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr5_-_34609337 | 0.06 |
ENSDART00000145792
|
hexb
|
hexosaminidase B (beta polypeptide) |
| chr22_+_39058269 | 0.06 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr15_-_2903475 | 0.05 |
ENSDART00000043226
|
guca1c
|
guanylate cyclase activator 1C |
| chr23_+_38953545 | 0.05 |
ENSDART00000184621
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr15_+_20543770 | 0.05 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr8_-_43689324 | 0.05 |
ENSDART00000159904
|
ep400
|
E1A binding protein p400 |
| chr23_-_9492266 | 0.05 |
ENSDART00000091852
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr16_+_16969060 | 0.05 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr21_+_39197628 | 0.05 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr24_+_37410403 | 0.05 |
ENSDART00000186282
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr20_+_29209767 | 0.04 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr9_-_53422084 | 0.04 |
ENSDART00000162661
|
gpc6b
|
glypican 6b |
| chr10_-_40964739 | 0.04 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr11_-_25734417 | 0.04 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr20_+_20731052 | 0.04 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr11_-_12158412 | 0.04 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr25_+_12494079 | 0.04 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr22_-_8720794 | 0.04 |
ENSDART00000191236
ENSDART00000185738 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr4_-_68868700 | 0.03 |
ENSDART00000187985
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr14_+_41409697 | 0.03 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr22_+_2533514 | 0.03 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr20_+_46620374 | 0.03 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr21_-_25756119 | 0.03 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr12_+_46608361 | 0.03 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr18_+_50089000 | 0.03 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr15_-_34468599 | 0.03 |
ENSDART00000192984
|
meox2a
|
mesenchyme homeobox 2a |
| chr2_-_17392799 | 0.03 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr14_+_9583588 | 0.03 |
ENSDART00000164101
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr11_-_26040594 | 0.03 |
ENSDART00000144115
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr20_+_18993452 | 0.03 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr4_+_37406676 | 0.03 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr19_-_10043142 | 0.03 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr6_+_9893554 | 0.03 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr21_+_7121942 | 0.03 |
ENSDART00000171603
|
mrps2
|
mitochondrial ribosomal protein S2 |
| chr23_+_21966447 | 0.03 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr1_-_35113974 | 0.03 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr3_-_31254979 | 0.02 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr13_-_49666615 | 0.02 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR+nr1i2_vdrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0035142 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.0 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:0031464 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |