Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
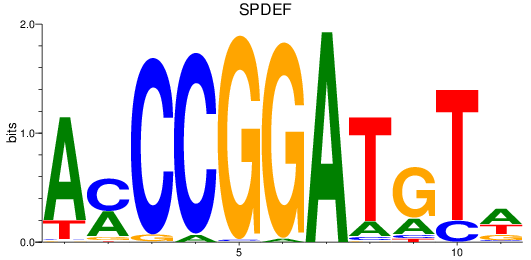
Results for SPDEF
Z-value: 1.87
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSDARG00000029930 | SAM pointed domain containing ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPDEF | dr11_v1_chr6_-_54290227_54290227 | -0.80 | 1.0e-02 | Click! |
Activity profile of SPDEF motif
Sorted Z-values of SPDEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_3398383 | 3.47 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr3_-_3413669 | 3.43 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr21_-_43636595 | 2.56 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr15_+_38299385 | 2.53 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr8_+_23726708 | 2.42 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr5_+_6854345 | 2.10 |
ENSDART00000066307
|
elac1
|
elaC ribonuclease Z 1 |
| chr15_+_38299563 | 2.06 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr12_-_9468618 | 2.05 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr18_+_6638974 | 1.95 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr8_+_23726244 | 1.95 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr1_+_49435017 | 1.88 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr18_+_6638726 | 1.85 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr23_-_33775145 | 1.77 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr6_+_4387150 | 1.74 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr15_+_34933552 | 1.72 |
ENSDART00000155368
|
zgc:66024
|
zgc:66024 |
| chr3_-_3372259 | 1.72 |
ENSDART00000140482
|
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr20_-_48898371 | 1.67 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr5_+_6854498 | 1.66 |
ENSDART00000148663
|
elac1
|
elaC ribonuclease Z 1 |
| chr2_+_32846602 | 1.62 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr2_+_51818039 | 1.61 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr20_-_48898560 | 1.53 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr2_+_30182431 | 1.51 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr8_+_48965767 | 1.49 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr15_+_25528290 | 1.48 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr25_+_8921425 | 1.48 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr5_-_26795438 | 1.48 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr6_+_3334392 | 1.47 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr8_+_48966165 | 1.43 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr22_-_5171362 | 1.41 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr20_+_13141408 | 1.38 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr17_-_7351488 | 1.37 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr8_-_33154677 | 1.37 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr21_-_30994577 | 1.36 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr15_-_43625549 | 1.34 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr7_-_71384391 | 1.34 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr8_+_17143501 | 1.34 |
ENSDART00000061758
|
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr17_+_8754020 | 1.32 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr1_+_24557414 | 1.29 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr7_+_35191220 | 1.28 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr12_+_46543572 | 1.28 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr20_-_39789036 | 1.27 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr8_+_30112655 | 1.27 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr1_-_40519340 | 1.27 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr8_-_14080534 | 1.25 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr13_+_31545530 | 1.22 |
ENSDART00000164590
ENSDART00000178460 ENSDART00000185503 |
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr17_+_8754426 | 1.21 |
ENSDART00000185519
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr22_+_8753092 | 1.20 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr18_+_3634652 | 1.18 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr13_+_31545812 | 1.17 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr1_+_46598502 | 1.15 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr24_-_41220538 | 1.15 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr22_+_8753365 | 1.14 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr20_-_23254876 | 1.14 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr20_+_27087539 | 1.14 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr5_+_41477954 | 1.14 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr6_-_18531760 | 1.13 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr24_-_7826489 | 1.13 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr15_-_34933560 | 1.10 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr3_+_36617024 | 1.09 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr12_+_29240124 | 1.09 |
ENSDART00000053761
ENSDART00000130172 |
bms1
|
BMS1 ribosome biogenesis factor |
| chr24_+_31459715 | 1.09 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr15_-_41311292 | 1.09 |
ENSDART00000133835
ENSDART00000085645 ENSDART00000134075 |
usp16
|
ubiquitin specific peptidase 16 |
| chr19_+_34230108 | 1.09 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr15_-_15230264 | 1.08 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr14_-_5407555 | 1.06 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr6_-_18531349 | 1.05 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr13_+_9559461 | 1.05 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr23_+_31979602 | 1.05 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr20_-_1268863 | 1.04 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr24_+_15020402 | 1.04 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr15_-_43284021 | 1.03 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr18_+_3169579 | 1.03 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr19_-_34742440 | 1.02 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr22_-_10752471 | 1.02 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr5_+_47863153 | 1.02 |
ENSDART00000051518
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr13_-_44782462 | 1.02 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr19_+_37118547 | 1.02 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr7_-_28058442 | 1.02 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr1_-_28950366 | 1.01 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr5_+_25084385 | 1.01 |
ENSDART00000134526
ENSDART00000111863 |
paxx
|
PAXX, non-homologous end joining factor |
| chr16_-_44673851 | 1.01 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr21_+_38033226 | 0.98 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr15_+_19884242 | 0.98 |
ENSDART00000154437
ENSDART00000054416 |
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr17_+_37253706 | 0.97 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr15_+_17100697 | 0.97 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_+_12292274 | 0.96 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr14_-_45558490 | 0.95 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
| chr8_+_47683539 | 0.93 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr6_-_54444929 | 0.92 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr6_-_54433995 | 0.92 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr13_-_33700461 | 0.91 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr9_-_41090048 | 0.90 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr6_+_3334710 | 0.90 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr1_+_41596099 | 0.90 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr12_-_14211293 | 0.90 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr3_-_27066451 | 0.90 |
ENSDART00000156228
ENSDART00000156311 |
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr8_-_19246342 | 0.89 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr20_+_29587995 | 0.89 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr7_+_35193832 | 0.88 |
ENSDART00000189002
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr6_-_29159888 | 0.88 |
ENSDART00000110288
|
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr1_-_44048798 | 0.87 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr21_+_38745094 | 0.87 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr11_+_24348425 | 0.86 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr14_-_23684814 | 0.85 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr13_-_42673978 | 0.85 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr21_-_11655010 | 0.85 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr16_+_38337783 | 0.84 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr6_-_34860574 | 0.84 |
ENSDART00000073957
|
slc35d1a
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1a |
| chr9_+_25853052 | 0.84 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr9_-_24218367 | 0.84 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr15_+_46313082 | 0.83 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr5_+_28497956 | 0.82 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr20_-_23946296 | 0.80 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr15_-_766015 | 0.79 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr14_-_5407118 | 0.79 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr18_-_3552414 | 0.79 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr21_-_14762944 | 0.79 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr21_-_40938382 | 0.79 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| chr23_+_4260458 | 0.79 |
ENSDART00000103747
|
srsf6a
|
serine/arginine-rich splicing factor 6a |
| chr24_+_17069420 | 0.79 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr3_-_36419641 | 0.78 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr17_-_31579715 | 0.78 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr13_+_14006118 | 0.76 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr2_+_3044992 | 0.76 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr5_-_35456269 | 0.76 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr12_-_31724198 | 0.76 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr7_-_29356084 | 0.75 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr24_-_13349802 | 0.75 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr3_+_59880317 | 0.75 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr13_-_18069421 | 0.74 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr19_+_19241372 | 0.74 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr15_-_6650993 | 0.74 |
ENSDART00000002922
|
atm
|
ATM serine/threonine kinase |
| chr8_-_1267247 | 0.73 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr15_-_25365319 | 0.73 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr18_-_18584839 | 0.73 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr19_-_32042105 | 0.72 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr14_+_16151636 | 0.72 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr8_+_39770162 | 0.72 |
ENSDART00000190677
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr20_-_33174899 | 0.70 |
ENSDART00000047834
|
nbas
|
neuroblastoma amplified sequence |
| chr11_+_25693395 | 0.70 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr24_-_41195068 | 0.70 |
ENSDART00000121592
|
acvr2ba
|
activin A receptor type 2Ba |
| chr24_+_19522094 | 0.69 |
ENSDART00000191042
ENSDART00000184714 ENSDART00000180782 |
sulf1
|
sulfatase 1 |
| chr11_-_16115804 | 0.68 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr9_+_12905591 | 0.68 |
ENSDART00000139377
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr1_+_58990121 | 0.68 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr16_-_17345377 | 0.67 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr2_-_51741711 | 0.67 |
ENSDART00000171301
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr11_+_44236183 | 0.67 |
ENSDART00000193470
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr11_-_28050559 | 0.67 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr3_-_30152836 | 0.67 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr3_-_30153242 | 0.67 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr21_+_27302752 | 0.65 |
ENSDART00000012855
|
sart1
|
SART1, U4/U6.U5 tri-snRNP-associated protein 1 |
| chr13_-_12494575 | 0.65 |
ENSDART00000137761
|
si:dkey-20i10.7
|
si:dkey-20i10.7 |
| chr3_-_27065477 | 0.64 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr15_-_25365570 | 0.64 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr8_-_4031121 | 0.63 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr6_+_41808673 | 0.62 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr7_-_66693712 | 0.62 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr5_-_13086616 | 0.62 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr15_+_17100412 | 0.62 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr6_+_37625787 | 0.61 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr7_-_50349430 | 0.61 |
ENSDART00000065864
|
ttc17
|
tetratricopeptide repeat domain 17 |
| chr15_-_2519640 | 0.61 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr8_-_1266181 | 0.60 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr1_+_47165842 | 0.60 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr7_+_38809241 | 0.60 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr3_-_34528306 | 0.60 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr25_-_24248000 | 0.60 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr5_+_29715040 | 0.60 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr7_-_13906409 | 0.59 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr24_-_13349464 | 0.59 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr4_-_22749553 | 0.59 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr2_+_16798652 | 0.59 |
ENSDART00000145778
ENSDART00000087120 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr4_-_20108833 | 0.59 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr16_-_6944927 | 0.59 |
ENSDART00000149620
|
pmvk
|
phosphomevalonate kinase |
| chr17_+_33375469 | 0.59 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr2_+_49457626 | 0.59 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr17_+_24684778 | 0.58 |
ENSDART00000146309
ENSDART00000082237 |
znf593
|
zinc finger protein 593 |
| chr1_+_46598764 | 0.58 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr5_-_32489796 | 0.57 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr14_-_10617923 | 0.57 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr16_+_33931032 | 0.57 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr2_+_4383061 | 0.57 |
ENSDART00000163986
|
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr20_+_715739 | 0.57 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr3_-_34136368 | 0.56 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr17_+_27456804 | 0.56 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr2_+_49457449 | 0.55 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr2_-_45510699 | 0.55 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr15_-_34668485 | 0.55 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr14_+_712115 | 0.55 |
ENSDART00000157494
ENSDART00000166516 ENSDART00000082017 ENSDART00000122374 ENSDART00000170203 |
ints10
|
integrator complex subunit 10 |
| chr2_+_46032678 | 0.54 |
ENSDART00000184382
ENSDART00000125971 |
gpc1b
|
glypican 1b |
| chr14_-_43616572 | 0.54 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr15_-_14642186 | 0.53 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr9_-_52490579 | 0.52 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr24_-_26369185 | 0.52 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr6_+_12482599 | 0.52 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr9_-_28255029 | 0.51 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr2_-_45510223 | 0.51 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr7_-_34062301 | 0.51 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr5_+_44806374 | 0.50 |
ENSDART00000184237
|
ctsla
|
cathepsin La |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPDEF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.8 | 3.8 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.4 | 1.3 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.4 | 1.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.3 | 1.0 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.3 | 1.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 1.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 1.4 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 0.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 1.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.7 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 2.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 5.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.8 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 2.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.7 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 1.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.2 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.5 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.0 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.5 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 3.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 1.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 2.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 5.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.5 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.8 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 1.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.1 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 1.1 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.7 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.6 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.8 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.5 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.3 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.7 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 0.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 3.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 3.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 0.7 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 1.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.6 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.0 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.7 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.8 | 3.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 2.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.3 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 1.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.3 | 1.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.3 | 1.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 2.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 3.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 1.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 0.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 0.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.7 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 3.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0043142 | DNA clamp loader activity(GO:0003689) single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 3.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 4.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |