Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
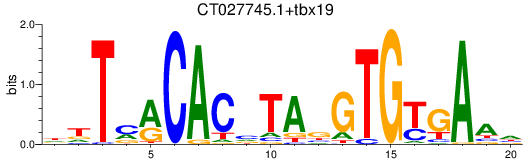
Results for CT027745.1+tbx19
Z-value: 1.71
Transcription factors associated with CT027745.1+tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx19
|
ENSDARG00000079187 | T-box transcription factor 19 |
|
CT027745.1
|
ENSDARG00000112623 | ENSDARG00000112623 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx19 | dr11_v1_chr6_+_9204099_9204099 | -0.36 | 3.5e-01 | Click! |
Activity profile of CT027745.1+tbx19 motif
Sorted Z-values of CT027745.1+tbx19 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_29025090 | 2.40 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr15_+_29024895 | 2.05 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr8_+_10862353 | 2.00 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr14_+_34490445 | 1.93 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr17_+_23311377 | 1.69 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr9_+_8365398 | 1.62 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr3_+_32416948 | 1.60 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_+_19764995 | 1.52 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr5_-_31773208 | 1.46 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr22_+_4443689 | 1.39 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr3_+_35510040 | 1.36 |
ENSDART00000162757
ENSDART00000182383 |
tnrc6a
|
trinucleotide repeat containing 6a |
| chr3_-_34724879 | 1.34 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr5_-_3960161 | 1.34 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr4_-_73447058 | 1.21 |
ENSDART00000172042
|
si:ch73-120g24.4
|
si:ch73-120g24.4 |
| chr10_-_33343244 | 1.18 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr2_-_37532772 | 1.15 |
ENSDART00000133951
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr15_-_25269028 | 1.15 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr17_-_33707589 | 1.14 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr25_+_18002368 | 1.12 |
ENSDART00000156000
|
tmtc3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr23_+_38957738 | 1.10 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr10_-_11840353 | 1.09 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr7_+_53152108 | 1.08 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
| chr5_+_29652513 | 1.05 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr9_-_55128839 | 1.02 |
ENSDART00000085135
|
tbl1x
|
transducin beta like 1 X-linked |
| chr5_-_18446483 | 1.02 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr22_+_35275468 | 1.02 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr4_-_29050235 | 0.99 |
ENSDART00000142951
|
si:dkey-23a23.2
|
si:dkey-23a23.2 |
| chr16_-_52879741 | 0.99 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr14_+_28486213 | 0.98 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr9_-_41025062 | 0.96 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr7_-_38570878 | 0.95 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr18_+_173603 | 0.95 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr5_-_23715861 | 0.95 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr9_-_41024438 | 0.94 |
ENSDART00000157398
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr23_+_38957472 | 0.93 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr19_-_17303500 | 0.90 |
ENSDART00000162355
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr9_+_27720428 | 0.88 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr9_-_27719998 | 0.87 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr11_-_24510995 | 0.86 |
ENSDART00000163489
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr2_+_1988036 | 0.86 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr1_+_218524 | 0.85 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr4_-_29050583 | 0.84 |
ENSDART00000102846
ENSDART00000181488 ENSDART00000181276 |
si:dkey-23a23.2
|
si:dkey-23a23.2 |
| chr13_-_44831072 | 0.84 |
ENSDART00000137275
ENSDART00000188781 |
si:dkeyp-2e4.6
|
si:dkeyp-2e4.6 |
| chr11_-_35970966 | 0.84 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr20_-_51831816 | 0.83 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr10_+_15603082 | 0.82 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr9_-_27720612 | 0.80 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr7_-_6368406 | 0.80 |
ENSDART00000172787
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr14_-_49859747 | 0.78 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr5_+_29652198 | 0.78 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr4_-_45136781 | 0.78 |
ENSDART00000141609
|
znf1046
|
zinc finger protein 1046 |
| chr5_-_69180587 | 0.77 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr5_-_66160415 | 0.76 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr5_+_1624359 | 0.75 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr22_-_7700179 | 0.75 |
ENSDART00000145047
|
si:ch211-59h6.1
|
si:ch211-59h6.1 |
| chr16_+_35595312 | 0.74 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr23_+_4777348 | 0.70 |
ENSDART00000171971
|
vgll4a
|
vestigial-like family member 4a |
| chr25_-_18002498 | 0.70 |
ENSDART00000158688
|
cep290
|
centrosomal protein 290 |
| chr5_-_37875636 | 0.69 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr2_-_31798717 | 0.69 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr12_+_10115964 | 0.68 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr3_+_9077907 | 0.68 |
ENSDART00000163056
|
znf985
|
zinc finger protein 985 |
| chr12_-_28956921 | 0.68 |
ENSDART00000142933
|
znf646
|
zinc finger protein 646 |
| chr10_+_9595575 | 0.67 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr9_-_29844596 | 0.67 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr23_-_31913231 | 0.67 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr5_+_67268110 | 0.67 |
ENSDART00000148116
|
si:ch211-110p13.9
|
si:ch211-110p13.9 |
| chr20_-_32188897 | 0.65 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr3_+_55105066 | 0.65 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr18_+_44750170 | 0.64 |
ENSDART00000132775
ENSDART00000187468 |
si:dkey-224e22.2
|
si:dkey-224e22.2 |
| chr2_-_31800521 | 0.64 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr4_+_69231190 | 0.63 |
ENSDART00000158735
|
zgc:173615
|
zgc:173615 |
| chr18_-_33979693 | 0.63 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr14_-_6527010 | 0.61 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr7_-_19693762 | 0.61 |
ENSDART00000162994
|
si:ch211-196p9.1
|
si:ch211-196p9.1 |
| chr23_-_32162810 | 0.60 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_-_24231139 | 0.60 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr13_+_8696825 | 0.59 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr17_+_34186632 | 0.59 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr4_-_37707809 | 0.59 |
ENSDART00000162606
|
znf1139
|
zinc finger protein 1139 |
| chr20_+_9124369 | 0.57 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr4_-_54109066 | 0.57 |
ENSDART00000129685
ENSDART00000165283 ENSDART00000169701 |
si:dkey-199m13.7
|
si:dkey-199m13.7 |
| chr17_-_45164291 | 0.57 |
ENSDART00000062109
|
noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr22_-_31020690 | 0.57 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr24_+_17145830 | 0.56 |
ENSDART00000066763
ENSDART00000148756 ENSDART00000149744 ENSDART00000121821 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr23_-_3721444 | 0.56 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr5_+_56277866 | 0.55 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr6_-_8392104 | 0.55 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr6_-_39631164 | 0.54 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr11_-_16115804 | 0.54 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr4_+_69863750 | 0.54 |
ENSDART00000170800
|
znf1117
|
zinc finger protein 1117 |
| chr23_-_900795 | 0.53 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr4_+_76353418 | 0.53 |
ENSDART00000171289
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr5_-_8765428 | 0.52 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr4_+_59425539 | 0.51 |
ENSDART00000127914
|
si:ch211-241n15.3
|
si:ch211-241n15.3 |
| chr17_-_20167206 | 0.51 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr13_-_9024004 | 0.51 |
ENSDART00000169564
|
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr4_-_63080760 | 0.51 |
ENSDART00000167958
|
CR450780.2
|
|
| chr25_-_10610961 | 0.50 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr17_+_21818093 | 0.50 |
ENSDART00000125335
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr4_-_65036768 | 0.50 |
ENSDART00000184455
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr4_-_48727718 | 0.50 |
ENSDART00000121499
|
znf1023
|
zinc finger protein 1023 |
| chr4_-_69230711 | 0.50 |
ENSDART00000163977
|
si:dkey-82i20.5
|
si:dkey-82i20.5 |
| chr4_+_42296225 | 0.50 |
ENSDART00000168471
|
znf1118
|
zinc finger protein 1118 |
| chr5_-_24543526 | 0.49 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr4_+_45195990 | 0.49 |
ENSDART00000135131
|
znf1021
|
zinc finger protein 1021 |
| chr6_+_40523370 | 0.48 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr16_+_7991274 | 0.48 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr3_-_55104310 | 0.48 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr17_-_50456604 | 0.47 |
ENSDART00000156110
|
si:ch211-235i11.7
|
si:ch211-235i11.7 |
| chr6_-_40651944 | 0.47 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr17_+_50389823 | 0.46 |
ENSDART00000154246
ENSDART00000153566 ENSDART00000153878 |
znf982
|
zinc finger protein 982 |
| chr4_+_63928858 | 0.46 |
ENSDART00000161958
|
si:dkey-179k24.2
|
si:dkey-179k24.2 |
| chr5_-_55201172 | 0.46 |
ENSDART00000134937
|
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr8_+_49975160 | 0.45 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr4_+_37239915 | 0.45 |
ENSDART00000146015
|
znf973
|
zinc finger protein 973 |
| chr19_+_32201991 | 0.45 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr2_-_3419890 | 0.43 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr5_+_9147497 | 0.43 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr15_-_19443997 | 0.42 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr11_+_24800156 | 0.42 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr7_-_15185811 | 0.42 |
ENSDART00000031049
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr3_+_40164129 | 0.41 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr4_+_33819759 | 0.41 |
ENSDART00000188892
ENSDART00000182514 ENSDART00000133511 ENSDART00000160632 ENSDART00000169630 ENSDART00000146157 ENSDART00000182964 |
znf1026
|
zinc finger protein 1026 |
| chr5_-_24542726 | 0.40 |
ENSDART00000182975
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr8_-_18228809 | 0.39 |
ENSDART00000110249
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr20_+_27096430 | 0.38 |
ENSDART00000153358
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr4_-_37236494 | 0.38 |
ENSDART00000165241
|
si:dkey-3p4.7
|
si:dkey-3p4.7 |
| chr25_-_18002937 | 0.38 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr25_+_27410352 | 0.37 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr17_-_50490475 | 0.37 |
ENSDART00000075159
|
zgc:113886
|
zgc:113886 |
| chr12_-_15567104 | 0.37 |
ENSDART00000053003
|
hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr4_+_50891961 | 0.34 |
ENSDART00000150271
|
si:ch211-208f21.2
|
si:ch211-208f21.2 |
| chr11_-_36446655 | 0.34 |
ENSDART00000179025
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr4_-_55321457 | 0.33 |
ENSDART00000190996
|
znf998
|
zinc finger protein 998 |
| chr5_+_1877464 | 0.33 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr6_-_40899618 | 0.33 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr4_+_35876704 | 0.32 |
ENSDART00000159084
|
si:dkey-176f19.7
|
si:dkey-176f19.7 |
| chr1_+_55662491 | 0.31 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr19_-_24745317 | 0.31 |
ENSDART00000142774
|
MYO1G
|
si:dkeyp-92c9.3 |
| chr4_+_60296027 | 0.30 |
ENSDART00000172479
|
si:dkey-248e17.5
|
si:dkey-248e17.5 |
| chr14_+_22132896 | 0.30 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr4_-_51044211 | 0.30 |
ENSDART00000168119
ENSDART00000168584 ENSDART00000193247 ENSDART00000130293 ENSDART00000185716 ENSDART00000162333 |
znf1026
|
zinc finger protein 1026 |
| chr20_+_38837238 | 0.29 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr1_-_43963441 | 0.29 |
ENSDART00000137646
|
odam
|
odontogenic, ameloblast associated |
| chr3_+_36972298 | 0.28 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr5_-_42059869 | 0.27 |
ENSDART00000193984
|
cenpv
|
centromere protein V |
| chr4_+_45195202 | 0.26 |
ENSDART00000150838
ENSDART00000187011 |
znf1021
BX649479.1
|
zinc finger protein 1021 |
| chr2_-_26563978 | 0.26 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr8_-_18229169 | 0.26 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr7_-_20158380 | 0.25 |
ENSDART00000142891
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr9_+_20853894 | 0.25 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr1_-_45614481 | 0.25 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr22_-_35006554 | 0.24 |
ENSDART00000138639
|
kcnip2
|
Kv channel interacting protein 2 |
| chr1_+_18811679 | 0.24 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr24_-_11908115 | 0.23 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr4_-_59094319 | 0.23 |
ENSDART00000162560
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr5_-_69923241 | 0.23 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr22_+_9523479 | 0.23 |
ENSDART00000189473
ENSDART00000143953 |
strip1
|
striatin interacting protein 1 |
| chr6_+_21992820 | 0.22 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr14_+_34501245 | 0.21 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr12_-_27031060 | 0.20 |
ENSDART00000076103
|
chmp2a
|
charged multivesicular body protein 2A |
| chr8_+_47817454 | 0.19 |
ENSDART00000144825
|
pimr185
|
Pim proto-oncogene, serine/threonine kinase, related 185 |
| chr10_+_36322894 | 0.19 |
ENSDART00000166299
|
or106-3
|
odorant receptor, family G, subfamily 106, member 3 |
| chr12_+_38929663 | 0.18 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr2_-_20788698 | 0.18 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr17_-_36483787 | 0.17 |
ENSDART00000173306
|
DCDC2C
|
si:dkey-25g12.4 |
| chr4_+_39055027 | 0.17 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr5_+_59392183 | 0.17 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr4_+_42295479 | 0.17 |
ENSDART00000162279
|
znf1118
|
zinc finger protein 1118 |
| chr22_-_8648144 | 0.15 |
ENSDART00000078241
ENSDART00000191246 |
zmp:0000001175
|
zmp:0000001175 |
| chr16_+_11573471 | 0.14 |
ENSDART00000145858
|
si:dkey-11o1.2
|
si:dkey-11o1.2 |
| chr17_-_8899323 | 0.13 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr4_+_52356485 | 0.13 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr21_-_45382112 | 0.13 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr11_+_12860438 | 0.13 |
ENSDART00000037474
|
zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr11_+_12860260 | 0.13 |
ENSDART00000190410
|
zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr18_-_33979422 | 0.12 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr18_+_45114392 | 0.11 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr13_-_37545487 | 0.10 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr9_-_54304684 | 0.10 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr5_-_32890807 | 0.10 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr4_-_56083825 | 0.10 |
ENSDART00000157710
|
znf986
|
zinc finger protein 986 |
| chr17_-_24890843 | 0.10 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr2_-_27315604 | 0.09 |
ENSDART00000185540
|
dsela
|
dermatan sulfate epimerase-like a |
| chr24_-_35534273 | 0.09 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr4_+_65057234 | 0.08 |
ENSDART00000162985
|
si:dkey-14o6.2
|
si:dkey-14o6.2 |
| chr6_+_46258866 | 0.08 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr15_+_15786160 | 0.08 |
ENSDART00000130670
ENSDART00000090939 |
tada2a
|
transcriptional adaptor 2A |
| chr21_-_40040178 | 0.08 |
ENSDART00000159741
|
BX248108.1
|
Danio rerio multidrug and toxin extrusion protein 1-like (mate4), mRNA. |
| chr2_-_27315438 | 0.07 |
ENSDART00000087687
|
dsela
|
dermatan sulfate epimerase-like a |
| chr1_-_43892349 | 0.06 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr20_+_18994059 | 0.05 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr16_+_33902006 | 0.04 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_+_19943803 | 0.03 |
ENSDART00000164661
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr5_-_42060121 | 0.03 |
ENSDART00000148021
ENSDART00000147407 |
cenpv
|
centromere protein V |
| chr7_+_17063761 | 0.03 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr4_+_69863019 | 0.02 |
ENSDART00000168400
|
znf1117
|
zinc finger protein 1117 |
| chr6_-_10698125 | 0.02 |
ENSDART00000151636
|
dnah9l
|
dynein, axonemal, heavy polypeptide 9 like |
| chr4_+_51160367 | 0.01 |
ENSDART00000189984
|
znf1141
|
zinc finger protein 1141 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CT027745.1+tbx19
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 1.7 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.5 | 1.4 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.3 | 1.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 0.9 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.9 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 1.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 2.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 1.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.6 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.8 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 1.7 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.4 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.6 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.8 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.7 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.6 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.0 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 1.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.4 | 1.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 1.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 4.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 2.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.2 | 0.7 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.6 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |