Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
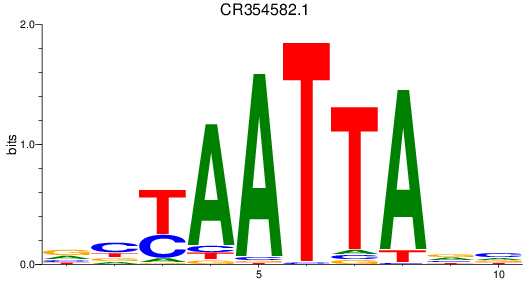
Results for CR354582.1
Z-value: 0.45
Transcription factors associated with CR354582.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CR354582.1
|
ENSDARG00000110598 | ENSDARG00000110598 |
Activity profile of CR354582.1 motif
Sorted Z-values of CR354582.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 0.91 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr9_-_35633827 | 0.69 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr18_-_40708537 | 0.52 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr2_-_15324837 | 0.51 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr24_+_1023839 | 0.51 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr11_-_44801968 | 0.44 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr20_-_23426339 | 0.42 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr12_-_14143344 | 0.39 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr10_-_34002185 | 0.39 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr10_-_21362320 | 0.38 |
ENSDART00000189789
|
avd
|
avidin |
| chr6_+_28208973 | 0.37 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr2_+_6253246 | 0.36 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_-_21362071 | 0.36 |
ENSDART00000125167
|
avd
|
avidin |
| chr11_+_18183220 | 0.35 |
ENSDART00000113468
|
LO018315.10
|
|
| chr2_+_50608099 | 0.35 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr8_-_23780334 | 0.35 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr12_-_33357655 | 0.35 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_-_8940499 | 0.34 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr10_+_6884627 | 0.34 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr23_+_2728095 | 0.33 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr8_+_45334255 | 0.31 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_-_42056137 | 0.30 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr11_+_30244356 | 0.29 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr11_-_1550709 | 0.28 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr10_-_32494499 | 0.26 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr13_+_38814521 | 0.25 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr1_-_18811517 | 0.25 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_-_21142550 | 0.24 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr10_-_32494304 | 0.24 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr22_+_28337429 | 0.23 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr10_+_6884123 | 0.23 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr19_-_30510259 | 0.23 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr22_+_28337204 | 0.23 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr22_+_4488454 | 0.23 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr9_+_50001746 | 0.22 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr6_+_9870192 | 0.21 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr19_-_5103141 | 0.21 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_-_18993110 | 0.20 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr10_-_11385155 | 0.20 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr20_-_29864390 | 0.20 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr3_-_50443607 | 0.20 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr11_+_31864921 | 0.20 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr22_-_21897203 | 0.19 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr7_+_22313533 | 0.19 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr17_-_200316 | 0.19 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr17_-_41798856 | 0.19 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr5_+_37903790 | 0.19 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr24_+_16547035 | 0.19 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_+_18811679 | 0.18 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr24_+_39518774 | 0.18 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr21_-_14174786 | 0.18 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr13_+_38817871 | 0.18 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr3_+_28860283 | 0.18 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr11_-_29768054 | 0.18 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr8_+_11425048 | 0.18 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr19_-_5103313 | 0.18 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr19_+_2631565 | 0.18 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr6_-_12172424 | 0.18 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr1_-_55248496 | 0.18 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr17_-_40956035 | 0.17 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr18_+_1837668 | 0.17 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr9_-_50001606 | 0.17 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr23_-_14990865 | 0.17 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr20_-_40755614 | 0.16 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr5_+_6954162 | 0.16 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr14_-_2933185 | 0.16 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr5_-_9625459 | 0.16 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr2_-_55298075 | 0.15 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr4_+_21129752 | 0.15 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_+_10339538 | 0.14 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr19_+_31585917 | 0.14 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr7_+_34592526 | 0.14 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr3_+_17933132 | 0.14 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr21_-_39177564 | 0.14 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr25_+_35891342 | 0.14 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr24_-_31843173 | 0.14 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr9_-_22057658 | 0.13 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr17_+_16046132 | 0.13 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_-_37813863 | 0.13 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr1_-_29139141 | 0.13 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr21_-_3853204 | 0.13 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_+_19933356 | 0.13 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr9_-_50000144 | 0.12 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_+_42471455 | 0.12 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr24_-_25144441 | 0.12 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_-_881725 | 0.12 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr15_-_9272328 | 0.12 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr13_-_31017960 | 0.12 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr25_-_13490744 | 0.12 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr18_-_43884044 | 0.12 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr4_+_9400012 | 0.12 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr4_+_13586689 | 0.12 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr10_+_25726694 | 0.12 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr19_+_390298 | 0.11 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr10_-_13343831 | 0.11 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr5_-_67629263 | 0.11 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_-_33945692 | 0.11 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr14_+_44804326 | 0.11 |
ENSDART00000079866
ENSDART00000172974 |
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr24_+_14581864 | 0.11 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr5_+_63302660 | 0.10 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr15_-_16177603 | 0.10 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr13_+_29462249 | 0.10 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr10_-_15963903 | 0.10 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr17_+_37227936 | 0.10 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr5_+_66433287 | 0.10 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr6_-_43283122 | 0.10 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr8_-_19467011 | 0.10 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr17_-_14966384 | 0.09 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr7_-_7764287 | 0.09 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr15_+_31344472 | 0.09 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr10_-_2971407 | 0.09 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr20_-_45060241 | 0.09 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr2_+_39021282 | 0.09 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr20_-_9462433 | 0.09 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr14_+_23717165 | 0.09 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_+_35257651 | 0.09 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr9_-_746317 | 0.09 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr14_+_25817628 | 0.09 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr21_-_32781612 | 0.08 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr7_+_30787903 | 0.08 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_-_23768234 | 0.08 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr5_-_68093169 | 0.08 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr14_+_8940326 | 0.08 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr8_-_25034411 | 0.08 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr20_+_29209767 | 0.08 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr9_+_22485343 | 0.08 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr14_+_34490445 | 0.08 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr2_+_44615000 | 0.08 |
ENSDART00000188826
ENSDART00000113232 |
yeats2
|
YEATS domain containing 2 |
| chr1_+_513986 | 0.08 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr19_+_42432625 | 0.08 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr18_+_8320165 | 0.08 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr19_+_9174166 | 0.07 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr22_-_16416882 | 0.07 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr20_-_9436521 | 0.07 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr15_-_18115540 | 0.07 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr24_+_13316737 | 0.07 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr17_+_21546993 | 0.07 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr10_-_35257458 | 0.07 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr21_-_27185915 | 0.07 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr7_-_72261721 | 0.07 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr24_+_40860320 | 0.07 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr12_+_30367371 | 0.07 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr12_+_30367079 | 0.07 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr24_-_34680956 | 0.07 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_-_12464412 | 0.06 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr10_-_26512993 | 0.06 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr23_+_40460333 | 0.06 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr18_-_15551360 | 0.06 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr18_+_2228737 | 0.06 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr12_+_48803098 | 0.06 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr19_-_19379084 | 0.06 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr10_-_26512742 | 0.06 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr6_-_55585423 | 0.06 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr19_+_43297546 | 0.06 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr23_+_33963619 | 0.05 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr6_-_40922971 | 0.05 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr5_-_15851953 | 0.05 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr12_+_22580579 | 0.05 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr10_-_26744131 | 0.05 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr13_-_42400647 | 0.05 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr15_+_47903864 | 0.05 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr6_+_40922572 | 0.05 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr25_-_25384045 | 0.05 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr24_-_2450597 | 0.04 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr20_+_37294112 | 0.04 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr24_-_16899406 | 0.04 |
ENSDART00000148753
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr25_-_10503043 | 0.04 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr14_+_40874608 | 0.04 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr3_+_13929860 | 0.04 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr6_+_52873822 | 0.04 |
ENSDART00000103138
|
or137-3
|
odorant receptor, family H, subfamily 137, member 3 |
| chr20_+_29209926 | 0.04 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_-_14126646 | 0.04 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr5_+_30624183 | 0.04 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr5_-_50992690 | 0.04 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr16_-_22930925 | 0.03 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr16_-_17347727 | 0.03 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr22_-_20166660 | 0.03 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr17_+_16046314 | 0.03 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_-_10586191 | 0.03 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr18_-_1185772 | 0.03 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr6_+_102506 | 0.03 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr11_+_7264457 | 0.03 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr22_+_20427170 | 0.03 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr19_-_13808630 | 0.03 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr19_-_32641725 | 0.03 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr9_+_41156818 | 0.03 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr17_+_3379673 | 0.03 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr16_+_17389116 | 0.03 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr11_+_18873619 | 0.02 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr22_-_5518117 | 0.02 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr2_-_32384683 | 0.02 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr19_-_20113696 | 0.02 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr24_-_4782052 | 0.02 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr15_-_34785594 | 0.02 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr3_+_13624815 | 0.02 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr25_-_6049339 | 0.02 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr25_-_13703826 | 0.02 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr21_-_31013817 | 0.02 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr12_-_19007834 | 0.02 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr19_-_32710922 | 0.01 |
ENSDART00000004034
|
hpca
|
hippocalcin |
Network of associatons between targets according to the STRING database.
First level regulatory network of CR354582.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.6 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |