Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
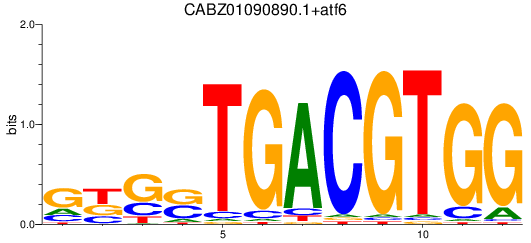
Results for CABZ01090890.1+atf6
Z-value: 0.87
Transcription factors associated with CABZ01090890.1+atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf6
|
ENSDARG00000012656 | activating transcription factor 6 |
|
CABZ01090890.1
|
ENSDARG00000101369 | ENSDARG00000101369 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf6 | dr11_v1_chr20_-_33909872_33909872 | 0.62 | 7.5e-02 | Click! |
Activity profile of CABZ01090890.1+atf6 motif
Sorted Z-values of CABZ01090890.1+atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_1487256 | 0.84 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr5_-_9540641 | 0.82 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr17_-_24879003 | 0.76 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr18_+_2153530 | 0.75 |
ENSDART00000158255
|
pygo1
|
pygopus family PHD finger 1 |
| chr3_-_2613990 | 0.62 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr11_+_440305 | 0.62 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr7_+_69449814 | 0.62 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr18_-_370286 | 0.61 |
ENSDART00000162633
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr9_+_54237100 | 0.59 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr9_+_24192370 | 0.59 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr5_-_48680580 | 0.59 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr15_-_43978141 | 0.58 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr11_-_42134968 | 0.57 |
ENSDART00000187115
|
FP325130.1
|
|
| chr1_+_36772348 | 0.57 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr5_+_44846280 | 0.57 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr20_+_41906960 | 0.56 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr18_+_8917766 | 0.56 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr5_-_30151815 | 0.55 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr21_+_20391978 | 0.55 |
ENSDART00000180817
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr1_+_36771954 | 0.55 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr21_-_3613702 | 0.55 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr6_-_29051773 | 0.54 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr3_-_46410387 | 0.54 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr23_-_25798099 | 0.53 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr21_+_10701834 | 0.53 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr17_+_50701748 | 0.52 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr3_+_37827373 | 0.52 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr8_+_1843135 | 0.51 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr21_+_10702031 | 0.51 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr8_-_44299247 | 0.50 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr23_+_20431140 | 0.49 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr3_-_26183699 | 0.49 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_+_20431388 | 0.48 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr13_+_9896845 | 0.48 |
ENSDART00000169076
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr3_-_55537096 | 0.47 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr10_-_28380919 | 0.45 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr6_-_1591002 | 0.45 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr9_+_22780901 | 0.45 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr25_-_37084032 | 0.44 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr9_+_21793565 | 0.44 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr12_+_2995887 | 0.44 |
ENSDART00000189499
ENSDART00000182597 ENSDART00000184918 ENSDART00000182292 |
lrrc45
|
leucine rich repeat containing 45 |
| chr15_+_17100697 | 0.43 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_+_31380495 | 0.42 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_-_27994679 | 0.42 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr12_+_23991276 | 0.42 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr1_-_36772147 | 0.42 |
ENSDART00000053369
|
prmt9
|
protein arginine methyltransferase 9 |
| chr15_+_44228917 | 0.42 |
ENSDART00000159630
|
CU929391.3
|
|
| chr20_-_50014936 | 0.41 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr15_-_3534705 | 0.40 |
ENSDART00000158150
|
cog6
|
component of oligomeric golgi complex 6 |
| chr18_+_20482369 | 0.40 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr10_+_9595575 | 0.40 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr6_+_58289335 | 0.40 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr20_+_22799857 | 0.39 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr3_+_36617024 | 0.39 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr16_-_22303130 | 0.39 |
ENSDART00000142181
|
si:dkey-92i15.4
|
si:dkey-92i15.4 |
| chr22_+_35275206 | 0.38 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr6_-_50685862 | 0.38 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr8_-_4100365 | 0.38 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr17_+_10090638 | 0.38 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr17_+_21102301 | 0.38 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr3_-_31845816 | 0.37 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr14_-_4044545 | 0.37 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr12_+_47081783 | 0.37 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr16_-_36064143 | 0.37 |
ENSDART00000158358
ENSDART00000182584 |
stk40
|
serine/threonine kinase 40 |
| chr18_+_619619 | 0.37 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr25_-_24247584 | 0.37 |
ENSDART00000046349
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr15_-_25153352 | 0.36 |
ENSDART00000078095
ENSDART00000122184 |
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr1_-_18592068 | 0.36 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr11_-_22916641 | 0.36 |
ENSDART00000080201
ENSDART00000154813 |
mdm4
|
MDM4, p53 regulator |
| chr3_-_47235997 | 0.36 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr4_+_68562464 | 0.36 |
ENSDART00000192954
|
BX548011.4
|
|
| chr14_-_45595711 | 0.36 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr9_-_27396404 | 0.36 |
ENSDART00000136412
ENSDART00000101401 |
tex30
|
testis expressed 30 |
| chr22_+_10713713 | 0.35 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr16_+_1228073 | 0.35 |
ENSDART00000109645
|
LO017852.1
|
|
| chr21_-_3844322 | 0.35 |
ENSDART00000166652
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr3_-_26184018 | 0.35 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr10_+_15603082 | 0.34 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr6_+_49901465 | 0.34 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr13_+_9896368 | 0.34 |
ENSDART00000137388
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr23_-_306796 | 0.34 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr10_+_2582254 | 0.34 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr19_-_1363759 | 0.33 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr9_+_33261330 | 0.33 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr7_+_13491452 | 0.33 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr3_+_41731527 | 0.33 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr4_+_23117557 | 0.33 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr17_+_584369 | 0.32 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr10_-_8672820 | 0.32 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr15_+_17100412 | 0.32 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_-_27099224 | 0.32 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr6_+_27275716 | 0.31 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr23_+_2703044 | 0.31 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr4_-_3145359 | 0.31 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr25_+_16080181 | 0.30 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr7_-_69352424 | 0.30 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr18_+_39487486 | 0.30 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr12_+_47162761 | 0.30 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr14_+_45970033 | 0.30 |
ENSDART00000047716
|
fermt3b
|
fermitin family member 3b |
| chr25_-_8138122 | 0.30 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr1_+_59146298 | 0.30 |
ENSDART00000191885
ENSDART00000152747 |
gpr108
|
G protein-coupled receptor 108 |
| chr22_+_32228882 | 0.30 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr6_-_11812224 | 0.30 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr17_+_17764979 | 0.29 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr25_+_16079913 | 0.29 |
ENSDART00000146350
|
far1
|
fatty acyl CoA reductase 1 |
| chr3_+_36616713 | 0.28 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr17_-_14671098 | 0.28 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr7_+_19835569 | 0.28 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr2_+_54387710 | 0.27 |
ENSDART00000127124
|
rab12
|
RAB12, member RAS oncogene family |
| chr1_+_10018466 | 0.27 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr10_-_15879569 | 0.26 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr15_+_24756860 | 0.26 |
ENSDART00000156424
ENSDART00000078035 |
cpda
|
carboxypeptidase D, a |
| chr22_+_17214389 | 0.25 |
ENSDART00000187083
ENSDART00000159468 ENSDART00000129109 ENSDART00000090107 |
nrd1b
|
nardilysin b (N-arginine dibasic convertase) |
| chr20_-_1191910 | 0.25 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr1_+_38142715 | 0.25 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr9_+_50038347 | 0.25 |
ENSDART00000164712
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr13_-_48431766 | 0.24 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr20_+_22799641 | 0.24 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr8_-_13315304 | 0.24 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr11_+_24758967 | 0.24 |
ENSDART00000005616
ENSDART00000133481 |
rnpep
|
arginyl aminopeptidase (aminopeptidase B) |
| chr6_+_59176470 | 0.24 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr23_+_7692042 | 0.23 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr15_-_43768776 | 0.23 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr4_-_19693978 | 0.23 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr12_+_31783066 | 0.23 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr19_+_4443285 | 0.23 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr3_+_22984098 | 0.23 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr7_-_44963154 | 0.23 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr18_-_46280578 | 0.23 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr8_-_37056211 | 0.22 |
ENSDART00000135155
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr8_-_65189 | 0.21 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr25_-_36263115 | 0.21 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr10_+_16036573 | 0.21 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr2_+_10063747 | 0.21 |
ENSDART00000143876
ENSDART00000014088 ENSDART00000134554 |
cmpk
|
cytidylate kinase |
| chr10_+_5689510 | 0.21 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_16036246 | 0.21 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr25_-_24248000 | 0.20 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr6_-_28943056 | 0.20 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr24_+_854877 | 0.20 |
ENSDART00000188408
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr25_+_21098675 | 0.19 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr23_+_13375359 | 0.19 |
ENSDART00000151947
|
ZNF512B
|
si:dkey-256k13.2 |
| chr13_-_25842074 | 0.19 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr22_+_24157807 | 0.18 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_-_54387550 | 0.18 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr18_-_11729 | 0.18 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr15_+_47746176 | 0.18 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr22_+_1786230 | 0.18 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr2_-_38225388 | 0.18 |
ENSDART00000146485
ENSDART00000128043 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr4_+_357810 | 0.18 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr9_-_8979468 | 0.18 |
ENSDART00000134646
|
ing1
|
inhibitor of growth family, member 1 |
| chr16_+_7985886 | 0.18 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr22_+_2751887 | 0.18 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr3_-_36750068 | 0.17 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr23_+_40655306 | 0.17 |
ENSDART00000141958
|
cdh24a
|
cadherin 24, type 2a |
| chr7_+_13995792 | 0.17 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr18_-_25855263 | 0.17 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr7_-_29164818 | 0.16 |
ENSDART00000052348
|
exosc6
|
exosome component 6 |
| chr13_+_36923052 | 0.16 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_+_17642731 | 0.16 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr17_-_49438873 | 0.16 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr17_+_27134806 | 0.16 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr25_+_18877675 | 0.16 |
ENSDART00000170605
|
armc10
|
armadillo repeat containing 10 |
| chr4_-_73488406 | 0.15 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr3_+_24094581 | 0.15 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr1_-_36771712 | 0.15 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr5_-_41804398 | 0.15 |
ENSDART00000134492
|
alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr19_+_40248697 | 0.14 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr25_+_21098990 | 0.14 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr8_+_49065348 | 0.14 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr17_+_53297822 | 0.14 |
ENSDART00000168297
|
ddx24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr19_-_10324182 | 0.13 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr18_+_50523870 | 0.13 |
ENSDART00000151593
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr13_-_22699024 | 0.13 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr13_-_46917098 | 0.13 |
ENSDART00000183453
|
slc29a1a
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1a |
| chr3_+_431208 | 0.13 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr2_+_51645164 | 0.13 |
ENSDART00000169600
|
abhd4
|
abhydrolase domain containing 4 |
| chr22_+_1462177 | 0.13 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr10_+_22510280 | 0.13 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr4_-_25515513 | 0.13 |
ENSDART00000142276
ENSDART00000044043 |
rbm17
|
RNA binding motif protein 17 |
| chr15_-_35212462 | 0.12 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr3_-_18384501 | 0.12 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr21_+_40417152 | 0.12 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr3_+_54047342 | 0.12 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr19_-_10324573 | 0.12 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr7_-_34927961 | 0.12 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T cells 3a |
| chr22_-_4989803 | 0.11 |
ENSDART00000181359
ENSDART00000125265 ENSDART00000028634 ENSDART00000183294 |
cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr19_+_31904836 | 0.11 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr11_-_3897067 | 0.11 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr12_+_33919502 | 0.11 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr4_-_25515154 | 0.11 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr4_+_16787040 | 0.10 |
ENSDART00000039027
|
golt1ba
|
golgi transport 1Ba |
| chr8_+_20863025 | 0.10 |
ENSDART00000015891
ENSDART00000187089 ENSDART00000146989 |
fzr1a
|
fizzy/cell division cycle 20 related 1a |
| chr21_-_31252131 | 0.10 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr4_-_16883051 | 0.10 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr12_+_46708920 | 0.10 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr11_-_8320868 | 0.10 |
ENSDART00000160570
|
si:ch211-247n2.1
|
si:ch211-247n2.1 |
| chr22_+_2801464 | 0.10 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr17_+_30448452 | 0.10 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr3_-_15889508 | 0.09 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr21_-_40413191 | 0.09 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01090890.1+atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.9 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.6 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.3 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.8 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.4 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 2.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.7 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0060415 | cardiac muscle tissue morphogenesis(GO:0055008) muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.6 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.0 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |