Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
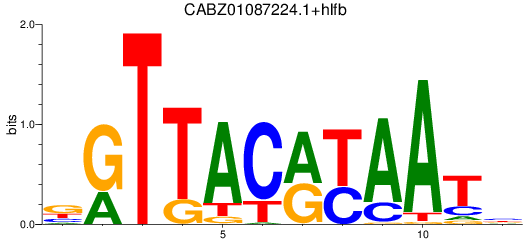
Results for CABZ01087224.1+hlfb
Z-value: 0.59
Transcription factors associated with CABZ01087224.1+hlfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlfb
|
ENSDARG00000061011 | HLF transcription factor, PAR bZIP family member b |
|
CABZ01087224.1
|
ENSDARG00000111269 | ENSDARG00000111269 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hlfb | dr11_v1_chr12_+_32159272_32159272 | -0.90 | 9.5e-04 | Click! |
| CABZ01087224.1 | dr11_v1_chr3_+_11568523_11568523 | 0.20 | 6.0e-01 | Click! |
Activity profile of CABZ01087224.1+hlfb motif
Sorted Z-values of CABZ01087224.1+hlfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_22974019 | 1.24 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr16_-_29387215 | 1.01 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr25_+_36292465 | 0.88 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr22_+_10606573 | 0.86 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr17_-_17759138 | 0.83 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr9_-_12888082 | 0.82 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr23_-_12453700 | 0.79 |
ENSDART00000091140
|
snx21
|
sorting nexin family member 21 |
| chr19_+_14109348 | 0.71 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr7_+_46020508 | 0.68 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr13_-_10620652 | 0.67 |
ENSDART00000135000
ENSDART00000191587 |
si:ch73-54n14.2
camkmt
|
si:ch73-54n14.2 calmodulin-lysine N-methyltransferase |
| chr22_+_10606863 | 0.64 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr3_+_7763114 | 0.63 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr20_-_33966148 | 0.61 |
ENSDART00000148111
|
selp
|
selectin P |
| chr7_+_15736230 | 0.57 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr4_+_5196469 | 0.57 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr3_+_41731527 | 0.57 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr20_-_35508805 | 0.55 |
ENSDART00000169538
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr20_+_1272526 | 0.54 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_+_44518636 | 0.54 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr5_-_30074332 | 0.54 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr23_-_18030399 | 0.53 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr14_-_30905288 | 0.52 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr13_-_25408387 | 0.52 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr1_-_354115 | 0.51 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr22_+_15973122 | 0.50 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr15_-_24960730 | 0.50 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr7_+_38380135 | 0.50 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr7_+_46019780 | 0.49 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr6_-_4228640 | 0.49 |
ENSDART00000162497
ENSDART00000179923 |
trak2
|
trafficking protein, kinesin binding 2 |
| chr10_+_6884123 | 0.48 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr16_+_47207691 | 0.48 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr15_-_1885247 | 0.48 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr2_-_55779927 | 0.47 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr5_-_33236637 | 0.46 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr18_+_25546227 | 0.46 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr8_+_247163 | 0.46 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr23_-_33361425 | 0.46 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr19_+_7549854 | 0.46 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr20_-_48877458 | 0.46 |
ENSDART00000163271
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr17_-_25831569 | 0.45 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr6_-_32411703 | 0.45 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr19_-_24757231 | 0.45 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr17_+_26722904 | 0.44 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr5_-_68782641 | 0.43 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr4_+_11690923 | 0.43 |
ENSDART00000150624
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr21_+_43178831 | 0.43 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr21_+_17051478 | 0.42 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr5_-_11809404 | 0.41 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr18_-_43866526 | 0.41 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr23_-_29812667 | 0.41 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr2_-_4070850 | 0.40 |
ENSDART00000159990
|
yme1l1b
|
YME1-like 1b |
| chr7_+_22853788 | 0.39 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr2_-_17115256 | 0.39 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_-_54395488 | 0.39 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr15_-_20412286 | 0.39 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr2_+_35603637 | 0.39 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr14_+_3507326 | 0.39 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr19_+_43715911 | 0.38 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_+_19687217 | 0.38 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr6_+_10333920 | 0.38 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr16_+_40954481 | 0.37 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr19_-_3056235 | 0.37 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr2_-_19234329 | 0.36 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr6_+_37754763 | 0.36 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr5_+_13870340 | 0.35 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr2_-_10563576 | 0.35 |
ENSDART00000185818
ENSDART00000190887 |
ccdc18
|
coiled-coil domain containing 18 |
| chr13_-_35472531 | 0.35 |
ENSDART00000057052
|
slx4ip
|
SLX4 interacting protein |
| chr12_-_34258384 | 0.35 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr3_-_26183699 | 0.35 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_-_30885250 | 0.35 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr20_-_33675676 | 0.35 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr2_+_16597011 | 0.35 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr24_-_26854032 | 0.35 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr1_-_45616242 | 0.35 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_+_8314451 | 0.34 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr15_+_12435975 | 0.34 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr23_-_35396845 | 0.34 |
ENSDART00000142038
ENSDART00000049373 ENSDART00000181978 ENSDART00000171357 |
cmtr1
|
cap methyltransferase 1 |
| chr18_+_8917766 | 0.34 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr7_+_38090515 | 0.34 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr9_+_38168012 | 0.34 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr9_-_29321625 | 0.34 |
ENSDART00000158689
ENSDART00000014047 ENSDART00000122602 |
pth2r
|
parathyroid hormone 2 receptor |
| chr21_+_18907102 | 0.33 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr5_+_27137473 | 0.33 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr21_-_32781612 | 0.33 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr8_+_26874924 | 0.33 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr20_-_23876291 | 0.33 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr2_-_10564019 | 0.33 |
ENSDART00000132167
|
ccdc18
|
coiled-coil domain containing 18 |
| chr18_+_17493859 | 0.33 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr7_+_73295890 | 0.32 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr3_+_25907266 | 0.32 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr2_-_37277626 | 0.32 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr14_-_30905963 | 0.32 |
ENSDART00000183543
ENSDART00000186441 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr11_+_43751263 | 0.32 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr1_+_12394205 | 0.31 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr2_+_9757453 | 0.31 |
ENSDART00000168972
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr18_-_14879135 | 0.31 |
ENSDART00000099701
|
selenoo1
|
selenoprotein O1 |
| chr18_-_15551360 | 0.31 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr20_-_20355577 | 0.31 |
ENSDART00000018500
|
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr6_-_7726849 | 0.31 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr3_-_40275096 | 0.31 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr3_-_26184018 | 0.31 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr17_+_25833947 | 0.30 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr5_-_14521500 | 0.30 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr8_-_53044300 | 0.30 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr23_+_19213472 | 0.30 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr19_+_42227400 | 0.30 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr9_+_21306902 | 0.30 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr10_+_23099890 | 0.30 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr3_+_14463941 | 0.30 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr17_+_17764979 | 0.30 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr18_-_39288894 | 0.30 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr23_-_25686894 | 0.30 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr20_+_1121458 | 0.30 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr16_-_30864824 | 0.30 |
ENSDART00000190444
ENSDART00000192231 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr19_+_15441022 | 0.30 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_+_34854562 | 0.29 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr13_-_25196758 | 0.29 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr20_-_38746889 | 0.29 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr21_+_1647990 | 0.29 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr16_+_35728992 | 0.29 |
ENSDART00000158442
|
map7d1a
|
MAP7 domain containing 1a |
| chr9_-_32300783 | 0.29 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr6_+_23810529 | 0.29 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr12_-_23365737 | 0.29 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr3_+_43373867 | 0.29 |
ENSDART00000159455
ENSDART00000172425 |
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr20_-_36617313 | 0.29 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr13_-_15142280 | 0.29 |
ENSDART00000163132
|
rab11fip5a
|
RAB11 family interacting protein 5a (class I) |
| chr23_+_9522781 | 0.29 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr9_-_5318873 | 0.29 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr24_-_23784701 | 0.29 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr1_-_58887610 | 0.28 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr8_-_4010887 | 0.28 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr8_+_26007988 | 0.28 |
ENSDART00000193948
ENSDART00000058100 |
xpc
|
xeroderma pigmentosum, complementation group C |
| chr19_-_30810328 | 0.28 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr6_+_36381709 | 0.28 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr4_-_27099224 | 0.28 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr1_+_19764995 | 0.28 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr3_+_33440615 | 0.28 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr8_+_12930216 | 0.28 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr8_-_25846188 | 0.28 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr10_-_2875735 | 0.28 |
ENSDART00000034555
|
ddx56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr15_+_24676905 | 0.27 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr20_+_474288 | 0.27 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr20_+_39250673 | 0.27 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr7_+_18176162 | 0.27 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr7_-_40959667 | 0.27 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr13_-_14929236 | 0.27 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr1_+_16600690 | 0.27 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr6_+_12326267 | 0.27 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr7_-_8961941 | 0.26 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr7_+_12950507 | 0.26 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr19_-_30811161 | 0.26 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr9_+_21268739 | 0.26 |
ENSDART00000186514
|
BX511129.3
|
|
| chr9_-_18743012 | 0.26 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr13_+_30035253 | 0.26 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr4_-_17353972 | 0.26 |
ENSDART00000041529
|
parpbp
|
PARP1 binding protein |
| chr7_+_22792895 | 0.26 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr21_+_38855551 | 0.25 |
ENSDART00000171977
|
ddx52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr6_+_18423402 | 0.25 |
ENSDART00000159747
|
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr2_+_30182431 | 0.25 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr25_-_12803723 | 0.25 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr19_+_46259619 | 0.25 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr14_-_30901602 | 0.25 |
ENSDART00000172925
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr22_-_17677947 | 0.25 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr4_+_5341592 | 0.25 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr9_-_27391908 | 0.25 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr9_-_9225980 | 0.25 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr18_+_5917625 | 0.25 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr12_+_48390715 | 0.25 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr24_+_26329018 | 0.25 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr9_-_13871935 | 0.24 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr12_+_46869271 | 0.24 |
ENSDART00000166560
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr13_-_42536642 | 0.24 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr11_+_24820542 | 0.24 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr24_+_32525146 | 0.24 |
ENSDART00000132417
ENSDART00000110185 |
yme1l1a
|
YME1-like 1a |
| chr18_+_21951551 | 0.24 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr15_+_21882419 | 0.24 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr9_+_27720428 | 0.24 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr12_+_32199651 | 0.24 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr16_-_35427060 | 0.24 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr21_-_32467099 | 0.24 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr9_+_22375779 | 0.24 |
ENSDART00000183956
|
dgkg
|
diacylglycerol kinase, gamma |
| chr6_+_8315050 | 0.24 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr8_+_7801060 | 0.23 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr1_+_37312580 | 0.23 |
ENSDART00000171340
|
fam193a
|
family with sequence similarity 193, member A |
| chr13_-_44836727 | 0.23 |
ENSDART00000144385
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr13_+_15816573 | 0.23 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr7_-_46019756 | 0.23 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr5_+_27421639 | 0.23 |
ENSDART00000146285
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr13_+_25396896 | 0.23 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr17_-_5610514 | 0.23 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr7_+_34297271 | 0.23 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_4931266 | 0.23 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr13_+_25397098 | 0.23 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr11_+_13176568 | 0.22 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr22_+_18816662 | 0.22 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr19_+_16016038 | 0.22 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr21_-_38618540 | 0.22 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr17_+_51744450 | 0.22 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr9_-_27720612 | 0.22 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr5_+_36439405 | 0.22 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01087224.1+hlfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.4 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.4 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.3 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 0.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.4 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.2 | GO:0070197 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.5 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.2 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.5 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.3 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.2 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.3 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.5 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.5 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.4 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0051883 | acute-phase response(GO:0006953) killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.6 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 0.4 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.4 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:2001270 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.4 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 1.2 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0051877 | chaperone-mediated protein complex assembly(GO:0051131) pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.5 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0010878 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.7 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 1.4 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 2.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.4 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.1 | 0.4 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.3 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.5 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 0.2 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.4 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.4 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.2 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.2 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0048018 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |