Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for CABZ01066694.1
Z-value: 0.43
Transcription factors associated with CABZ01066694.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CABZ01066694.1
|
ENSDARG00000110178 | ENSDARG00000110178 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01066694.1 | dr11_v1_chr6_+_168962_168962 | 0.04 | 9.3e-01 | Click! |
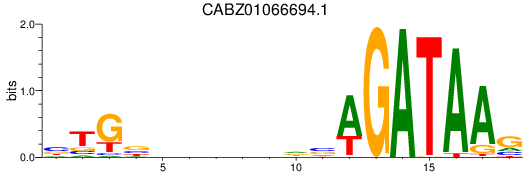
Activity profile of CABZ01066694.1 motif
Sorted Z-values of CABZ01066694.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20091964 | 0.64 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr9_-_22205682 | 0.44 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr8_+_31872992 | 0.40 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr16_-_45069882 | 0.40 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr10_-_17484762 | 0.37 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr15_+_32711663 | 0.34 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr8_+_21353878 | 0.31 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr11_+_41981959 | 0.30 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr11_-_45185792 | 0.29 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr2_+_16160906 | 0.29 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr15_+_29088426 | 0.28 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr21_-_11834452 | 0.28 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr14_+_9462726 | 0.27 |
ENSDART00000063559
|
rell2
|
RELT-like 2 |
| chr12_+_18524953 | 0.26 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr20_-_26066020 | 0.25 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr16_-_17300030 | 0.24 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr15_+_36115955 | 0.24 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr23_-_5685023 | 0.23 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr2_-_32768951 | 0.20 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr23_+_4689626 | 0.18 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr1_+_25585 | 0.18 |
ENSDART00000161139
|
CU651657.1
|
|
| chr21_-_44565236 | 0.18 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr2_-_2020044 | 0.18 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr23_+_22658700 | 0.17 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr12_-_20373058 | 0.17 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr3_+_33341640 | 0.17 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr5_+_67971627 | 0.17 |
ENSDART00000144879
|
mtif3
|
mitochondrial translational initiation factor 3 |
| chr5_+_37840914 | 0.16 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr17_+_8175998 | 0.16 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr20_+_33981946 | 0.16 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr2_-_54054225 | 0.16 |
ENSDART00000167239
|
CABZ01050249.1
|
|
| chr1_-_23274038 | 0.15 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr16_-_24668620 | 0.15 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr14_+_34514336 | 0.15 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr4_+_76692834 | 0.15 |
ENSDART00000064315
ENSDART00000137653 |
ms4a17a.1
|
membrane-spanning 4-domains, subfamily A, member 17A.1 |
| chr23_-_24488696 | 0.14 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr15_-_23522653 | 0.14 |
ENSDART00000144685
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr22_-_30935510 | 0.13 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr24_+_18286427 | 0.13 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr15_-_39969988 | 0.13 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr25_+_6186823 | 0.13 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr25_-_32311048 | 0.13 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr19_-_35733401 | 0.12 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
| chr16_+_29630965 | 0.12 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr4_+_27100531 | 0.12 |
ENSDART00000115200
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr7_+_19502744 | 0.11 |
ENSDART00000171258
ENSDART00000173691 |
si:ch211-212k18.9
|
si:ch211-212k18.9 |
| chr20_+_6590220 | 0.11 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr10_+_17936441 | 0.11 |
ENSDART00000146489
ENSDART00000064866 |
prkab1a
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, a |
| chr16_-_30931651 | 0.11 |
ENSDART00000132643
|
slc45a4
|
solute carrier family 45, member 4 |
| chr6_-_32349153 | 0.10 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr21_-_25756119 | 0.10 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr4_+_69619 | 0.10 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr19_-_10920577 | 0.09 |
ENSDART00000165630
|
ecm1a
|
extracellular matrix protein 1a |
| chr20_-_22378401 | 0.09 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr12_+_7497882 | 0.09 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr5_+_9246458 | 0.09 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr23_+_28377360 | 0.08 |
ENSDART00000014983
ENSDART00000128831 ENSDART00000135178 ENSDART00000138621 |
zgc:153867
|
zgc:153867 |
| chr3_+_32698424 | 0.08 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr6_-_42336987 | 0.08 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr3_-_31019715 | 0.08 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr4_+_39742119 | 0.08 |
ENSDART00000176004
|
CR749167.2
|
|
| chr21_+_198269 | 0.08 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr6_-_42377307 | 0.08 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr9_+_41156818 | 0.08 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr18_+_20566817 | 0.08 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr11_+_25101220 | 0.08 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr25_-_27735290 | 0.08 |
ENSDART00000140663
|
zgc:153935
|
zgc:153935 |
| chr20_+_27194833 | 0.07 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr7_+_36898622 | 0.07 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr15_+_34592215 | 0.07 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr5_+_9246018 | 0.07 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr7_+_26138240 | 0.07 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr15_-_37719015 | 0.07 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr17_+_44780166 | 0.06 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr4_-_1839352 | 0.06 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr15_+_6786757 | 0.06 |
ENSDART00000013278
|
sirt2
|
sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) |
| chr8_-_4586696 | 0.06 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr4_+_59245846 | 0.06 |
ENSDART00000104921
|
si:dkey-105i14.1
|
si:dkey-105i14.1 |
| chr5_-_7829657 | 0.06 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr16_-_30931468 | 0.06 |
ENSDART00000187838
|
slc45a4
|
solute carrier family 45, member 4 |
| chr24_-_39121900 | 0.05 |
ENSDART00000020282
|
CU929145.1
|
|
| chr5_-_37209481 | 0.05 |
ENSDART00000158614
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr20_-_1439256 | 0.05 |
ENSDART00000002928
|
themis
|
thymocyte selection associated |
| chr9_+_41156287 | 0.05 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr15_-_21837207 | 0.05 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr6_+_3334710 | 0.05 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr22_+_9981079 | 0.05 |
ENSDART00000124236
|
BX539307.2
|
|
| chr10_+_21677058 | 0.05 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr7_-_5191797 | 0.05 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr10_+_41199660 | 0.05 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr5_-_30088070 | 0.04 |
ENSDART00000078115
ENSDART00000134209 |
sdhda
|
succinate dehydrogenase complex, subunit D, integral membrane protein a |
| chr18_-_39473055 | 0.04 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr7_+_9922607 | 0.04 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr5_-_7897316 | 0.04 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr10_+_38643304 | 0.04 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr3_-_11203689 | 0.04 |
ENSDART00000166328
|
BX323882.1
|
|
| chr2_+_47927026 | 0.04 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr14_+_21783229 | 0.04 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr2_-_35103069 | 0.04 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr16_+_54829293 | 0.04 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr4_+_37936249 | 0.04 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr6_+_28877306 | 0.04 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr12_+_34732262 | 0.04 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr6_-_49078263 | 0.04 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr4_-_1839192 | 0.03 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr3_-_7940522 | 0.03 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr15_+_36054864 | 0.03 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr10_+_40633990 | 0.03 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr16_+_54829574 | 0.03 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr7_-_50883433 | 0.03 |
ENSDART00000174314
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr25_+_12849609 | 0.02 |
ENSDART00000168144
|
ccl33.2
|
chemokine (C-C motif) ligand 33, duplicate 2 |
| chr25_-_16742438 | 0.02 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr25_+_13321307 | 0.02 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr10_+_21584195 | 0.02 |
ENSDART00000100599
|
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr6_-_36182115 | 0.02 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr10_-_31220558 | 0.02 |
ENSDART00000134866
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr18_+_33340868 | 0.02 |
ENSDART00000099139
|
v2rh14
|
vomeronasal 2 receptor, h14 |
| chr5_+_42388464 | 0.02 |
ENSDART00000191596
|
BX548073.13
|
|
| chr15_+_35253247 | 0.02 |
ENSDART00000066737
|
zgc:158328
|
zgc:158328 |
| chr2_+_49713592 | 0.02 |
ENSDART00000189624
|
BX323861.3
|
|
| chr16_-_50157187 | 0.02 |
ENSDART00000155162
|
nek10
|
NIMA-related kinase 10 |
| chr24_-_6678640 | 0.02 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr7_-_52417060 | 0.02 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr7_+_8324506 | 0.02 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr21_-_12030654 | 0.02 |
ENSDART00000139145
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr4_-_11132617 | 0.02 |
ENSDART00000150250
|
si:dkey-21h14.9
|
si:dkey-21h14.9 |
| chr15_+_16886196 | 0.02 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr23_+_32406009 | 0.01 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr13_+_42011287 | 0.01 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr3_-_37699992 | 0.01 |
ENSDART00000151193
|
gpatch8
|
G patch domain containing 8 |
| chr3_-_58205045 | 0.01 |
ENSDART00000130740
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr2_-_9527129 | 0.01 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr15_+_404891 | 0.01 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr14_+_17197132 | 0.01 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr1_+_2025085 | 0.01 |
ENSDART00000008332
|
slc5a3a
|
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3a |
| chr3_-_26183699 | 0.01 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr12_+_18445604 | 0.01 |
ENSDART00000078860
|
noxo1b
|
NADPH oxidase organizer 1b |
| chr15_+_45563491 | 0.01 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr14_+_21783400 | 0.01 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr9_+_52613820 | 0.01 |
ENSDART00000168753
ENSDART00000165580 ENSDART00000164769 |
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr1_+_26622214 | 0.01 |
ENSDART00000182040
ENSDART00000152643 |
coro2a
|
coronin, actin binding protein, 2A |
| chr10_+_40583930 | 0.01 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr4_+_1722899 | 0.01 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr22_+_2680895 | 0.01 |
ENSDART00000082245
|
zgc:194221
|
zgc:194221 |
| chr5_+_38200807 | 0.01 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr17_-_50301313 | 0.01 |
ENSDART00000125772
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr5_+_43782267 | 0.01 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr18_-_32709297 | 0.01 |
ENSDART00000163311
ENSDART00000158288 |
olfcg8
|
olfactory receptor C family, g8 |
| chr18_+_34478959 | 0.01 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr7_-_68419830 | 0.01 |
ENSDART00000191606
|
CU468723.2
|
|
| chr13_+_30169681 | 0.00 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr21_-_40040178 | 0.00 |
ENSDART00000159741
|
BX248108.1
|
Danio rerio multidrug and toxin extrusion protein 1-like (mate4), mRNA. |
| chr22_+_7923713 | 0.00 |
ENSDART00000167210
|
CABZ01034691.1
|
|
| chr4_-_75681004 | 0.00 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr4_-_11122638 | 0.00 |
ENSDART00000150600
|
si:dkey-21h14.10
|
si:dkey-21h14.10 |
| chr15_+_45563656 | 0.00 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr5_+_30520249 | 0.00 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr15_+_17621375 | 0.00 |
ENSDART00000158352
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01066694.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.2 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) positive regulation of DNA binding(GO:0043388) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |