Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
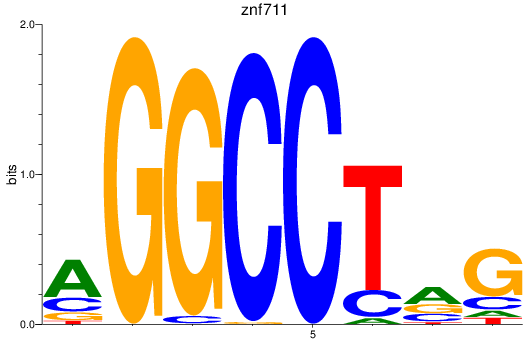
Results for znf711
Z-value: 2.64
Transcription factors associated with znf711
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf711
|
ENSDARG00000071868 | Danio rerio zinc finger protein 711 (znf711), transcript variant 1, mRNA. |
|
znf711
|
ENSDARG00000111963 | zinc finger protein 711 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf711 | dr11_v1_chr14_-_12020653_12020653 | 0.53 | 2.1e-02 | Click! |
Activity profile of znf711 motif
Sorted Z-values of znf711 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_23768898 | 11.52 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr7_-_48667056 | 5.79 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr13_+_45967179 | 5.15 |
ENSDART00000074499
|
olig4
|
oligodendrocyte transcription factor 4 |
| chr24_+_17005647 | 4.94 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr23_-_9859989 | 4.44 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr23_-_3758637 | 4.35 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr22_-_10541372 | 4.31 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr1_-_33557915 | 4.24 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr3_+_23743139 | 4.03 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr16_+_25163443 | 3.97 |
ENSDART00000105514
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr3_+_17653784 | 3.95 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr20_-_24183333 | 3.92 |
ENSDART00000025862
ENSDART00000153075 |
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr5_-_18007077 | 3.91 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr25_-_26736088 | 3.88 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr22_+_18349794 | 3.62 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr3_+_54581987 | 3.60 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr11_-_39105253 | 3.52 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr23_-_10175898 | 3.48 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr3_+_23742868 | 3.43 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr13_+_29778610 | 3.42 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr22_-_10541712 | 3.29 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr7_+_24393678 | 3.27 |
ENSDART00000188690
|
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr4_-_5831522 | 3.22 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr2_-_38080075 | 3.19 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr3_+_34149337 | 3.19 |
ENSDART00000006091
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr7_-_6279138 | 3.18 |
ENSDART00000173299
|
si:ch211-220f21.2
|
si:ch211-220f21.2 |
| chr14_+_31493306 | 3.12 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr2_+_3044992 | 3.11 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr2_+_16798923 | 3.10 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr14_+_41887070 | 3.08 |
ENSDART00000173264
|
slc7a11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr25_+_34845115 | 3.05 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr16_-_25680666 | 3.03 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr3_+_43086548 | 3.02 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr3_+_23752150 | 3.02 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr20_+_1329509 | 3.00 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr11_-_18107447 | 2.99 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr9_+_21306902 | 2.99 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr4_+_6032640 | 2.99 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr22_-_22242884 | 2.98 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr23_+_28378543 | 2.97 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr17_-_44247707 | 2.96 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr11_-_22599584 | 2.95 |
ENSDART00000014062
|
myog
|
myogenin |
| chr4_-_5831036 | 2.94 |
ENSDART00000166232
|
foxm1
|
forkhead box M1 |
| chr1_-_54972170 | 2.93 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr1_-_28885919 | 2.92 |
ENSDART00000152182
|
poglut1
|
protein O-glucosyltransferase 1 |
| chr19_+_48019070 | 2.88 |
ENSDART00000158269
|
UBE2M
|
si:ch1073-205c8.3 |
| chr8_-_1051438 | 2.85 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr16_+_31921812 | 2.84 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr5_-_23464970 | 2.84 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr23_+_35650771 | 2.81 |
ENSDART00000005158
|
ccnt1
|
cyclin T1 |
| chr2_-_37134169 | 2.80 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr1_-_23370395 | 2.80 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr3_-_5644028 | 2.80 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr15_+_24549054 | 2.77 |
ENSDART00000155900
|
phf12b
|
PHD finger protein 12b |
| chr3_+_18807524 | 2.76 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr17_-_42213285 | 2.76 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr14_+_31493119 | 2.76 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr20_-_24182689 | 2.74 |
ENSDART00000171184
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr11_-_3860534 | 2.71 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr11_-_18017918 | 2.70 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr16_+_25066880 | 2.70 |
ENSDART00000154084
|
im:7147486
|
im:7147486 |
| chr18_-_21748808 | 2.61 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr1_+_52792439 | 2.61 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr6_+_28877306 | 2.60 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr10_-_35186310 | 2.59 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr10_-_27197044 | 2.56 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr20_+_29209615 | 2.54 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr24_+_17140938 | 2.50 |
ENSDART00000149134
|
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr20_-_9199721 | 2.49 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr4_+_9177997 | 2.48 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr25_-_17378881 | 2.46 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr11_-_127785 | 2.46 |
ENSDART00000162658
|
ARFGAP1
|
ADP ribosylation factor GTPase activating protein 1 |
| chr13_+_21870269 | 2.43 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr19_+_31585917 | 2.42 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr16_+_28728347 | 2.42 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr9_-_32300611 | 2.40 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr1_-_19079957 | 2.40 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr23_-_43718067 | 2.40 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr3_+_32410746 | 2.38 |
ENSDART00000025496
|
rras
|
RAS related |
| chr15_-_4616816 | 2.37 |
ENSDART00000160191
ENSDART00000161721 ENSDART00000144949 |
eif4h
|
eukaryotic translation initiation factor 4h |
| chr8_+_26372115 | 2.37 |
ENSDART00000053460
|
nat6
|
N-acetyltransferase 6 |
| chr23_-_4704938 | 2.37 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr23_-_43486714 | 2.37 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr4_-_2059233 | 2.36 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr13_+_51710725 | 2.34 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr1_-_54971968 | 2.31 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr8_-_31416403 | 2.29 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr25_+_26901149 | 2.28 |
ENSDART00000153839
|
znf800a
|
zinc finger protein 800a |
| chr16_+_31922065 | 2.27 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr22_-_10752471 | 2.27 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr1_-_45580787 | 2.25 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_55183088 | 2.25 |
ENSDART00000100619
|
LUC7L2
|
zgc:158803 |
| chr5_+_30520249 | 2.25 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr3_+_22984098 | 2.23 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr20_-_18535502 | 2.23 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr18_-_46354269 | 2.23 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr23_-_9864166 | 2.21 |
ENSDART00000124510
ENSDART00000187934 |
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr24_+_21514283 | 2.20 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr25_-_8513255 | 2.20 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr1_+_59538755 | 2.20 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr7_+_72012397 | 2.19 |
ENSDART00000011303
|
ywhae2
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 |
| chr25_-_11026907 | 2.19 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr20_+_34770197 | 2.18 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr16_-_46578523 | 2.16 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr17_-_42213822 | 2.15 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr13_-_37600600 | 2.15 |
ENSDART00000003888
|
snapc1b
|
small nuclear RNA activating complex, polypeptide 1b |
| chr23_+_28378006 | 2.13 |
ENSDART00000188915
|
zgc:153867
|
zgc:153867 |
| chr18_-_15610856 | 2.12 |
ENSDART00000099849
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr18_+_44631789 | 2.12 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr19_+_48018802 | 2.11 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr18_-_24988645 | 2.07 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr24_+_4434117 | 2.06 |
ENSDART00000184349
|
GJD4
|
gap junction protein delta 4 |
| chr17_-_10838434 | 2.04 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr21_-_3007412 | 2.04 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr5_+_45000597 | 2.02 |
ENSDART00000051146
|
dmrt3a
|
doublesex and mab-3 related transcription factor 3a |
| chr13_-_44738574 | 2.01 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr25_-_3590923 | 1.99 |
ENSDART00000164149
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr23_-_29858087 | 1.97 |
ENSDART00000109506
|
tmem201
|
transmembrane protein 201 |
| chr7_+_22747915 | 1.97 |
ENSDART00000173609
|
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr1_-_57172294 | 1.95 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr25_+_17313568 | 1.95 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr23_-_16737161 | 1.94 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr3_+_50201240 | 1.91 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr9_-_33107237 | 1.91 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr7_+_20260172 | 1.91 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr20_+_26939742 | 1.90 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr13_+_11829072 | 1.89 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr16_+_20055878 | 1.89 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr4_-_9191220 | 1.86 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr6_-_39893501 | 1.86 |
ENSDART00000141611
ENSDART00000135631 ENSDART00000077662 ENSDART00000130613 |
myl6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr21_-_11970199 | 1.85 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_+_49654588 | 1.84 |
ENSDART00000025451
ENSDART00000141934 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr17_+_42027969 | 1.82 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr6_-_138392 | 1.82 |
ENSDART00000148974
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr13_-_36535128 | 1.81 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr8_+_12951155 | 1.80 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr24_-_7957581 | 1.80 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr7_-_66126628 | 1.80 |
ENSDART00000184492
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr17_+_4325693 | 1.80 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr16_-_39477746 | 1.79 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_-_72390259 | 1.79 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr16_-_31621719 | 1.78 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20 like 1 |
| chr11_+_6010177 | 1.77 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr16_-_26676685 | 1.76 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_-_55224 | 1.76 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr18_-_17147803 | 1.76 |
ENSDART00000187986
ENSDART00000169080 |
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr7_-_18508815 | 1.75 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr5_-_30516646 | 1.74 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr7_+_40081630 | 1.74 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr2_-_42173834 | 1.74 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_-_40529782 | 1.72 |
ENSDART00000055194
ENSDART00000141737 |
actb2
|
actin, beta 2 |
| chr9_-_29036263 | 1.72 |
ENSDART00000160393
|
tmem177
|
transmembrane protein 177 |
| chr15_-_35960250 | 1.71 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr6_+_32326074 | 1.71 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr19_+_31585341 | 1.70 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr1_-_35924495 | 1.70 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr16_+_16529748 | 1.69 |
ENSDART00000029579
|
ccdc12
|
coiled-coil domain containing 12 |
| chr13_+_32148338 | 1.68 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr16_+_20056030 | 1.67 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr13_-_23007813 | 1.67 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr5_-_1963498 | 1.67 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr25_-_21280584 | 1.67 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr1_+_8508753 | 1.66 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr7_-_59564011 | 1.62 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr25_+_30460378 | 1.62 |
ENSDART00000016310
|
banp
|
BTG3 associated nuclear protein |
| chr2_-_2813259 | 1.62 |
ENSDART00000032540
|
usp14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr18_-_50839033 | 1.61 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr4_-_35899188 | 1.61 |
ENSDART00000167093
|
zgc:174653
|
zgc:174653 |
| chr20_+_29209767 | 1.61 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_+_24280380 | 1.61 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr13_-_36534380 | 1.58 |
ENSDART00000177857
ENSDART00000181046 ENSDART00000187243 |
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr7_+_41146560 | 1.56 |
ENSDART00000143285
ENSDART00000173852 ENSDART00000174003 ENSDART00000038487 ENSDART00000173463 ENSDART00000166448 ENSDART00000052274 |
puf60b
|
poly-U binding splicing factor b |
| chr18_-_17147600 | 1.56 |
ENSDART00000141873
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr7_-_52531252 | 1.55 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr14_+_21686207 | 1.55 |
ENSDART00000034438
|
ran
|
RAN, member RAS oncogene family |
| chr17_-_44249538 | 1.55 |
ENSDART00000008816
|
otx2b
|
orthodenticle homeobox 2b |
| chr1_+_21731382 | 1.55 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr3_+_27607268 | 1.53 |
ENSDART00000024453
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr23_+_43718115 | 1.52 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr11_-_5953636 | 1.52 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr7_-_41915312 | 1.51 |
ENSDART00000159869
|
dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr3_-_15496295 | 1.49 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr24_-_26945390 | 1.49 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr9_+_8898024 | 1.49 |
ENSDART00000134954
ENSDART00000143671 ENSDART00000147098 ENSDART00000179969 ENSDART00000111214 ENSDART00000140232 ENSDART00000139687 ENSDART00000132443 ENSDART00000130208 |
naxd
|
NAD(P)HX dehydratase |
| chr9_-_25181137 | 1.48 |
ENSDART00000192624
|
esd
|
esterase D/formylglutathione hydrolase |
| chr5_-_12219572 | 1.47 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr23_+_24926407 | 1.47 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr3_+_32631372 | 1.47 |
ENSDART00000129339
|
srrm2
|
serine/arginine repetitive matrix 2 |
| chr5_-_33607062 | 1.46 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr24_+_36239617 | 1.44 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr2_-_3044947 | 1.43 |
ENSDART00000192642
|
guk1a
|
guanylate kinase 1a |
| chr13_+_11876437 | 1.42 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr25_+_5249513 | 1.40 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr9_-_25366541 | 1.40 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr4_+_18489207 | 1.39 |
ENSDART00000135276
|
si:dkey-202b22.5
|
si:dkey-202b22.5 |
| chr17_+_30751462 | 1.38 |
ENSDART00000154184
|
ldah
|
lipid droplet associated hydrolase |
| chr22_+_10752787 | 1.38 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_-_15496551 | 1.37 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr6_-_42369186 | 1.36 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr22_-_4769140 | 1.36 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf711
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.5 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 1.6 | 4.9 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.3 | 5.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.9 | 2.7 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.9 | 5.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.8 | 5.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.8 | 3.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.8 | 2.4 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.7 | 2.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.6 | 2.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.6 | 2.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.6 | 1.7 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.6 | 5.0 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.5 | 2.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 5.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.5 | 2.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.5 | 1.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.5 | 1.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 1.9 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.5 | 1.9 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.4 | 6.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.4 | 1.7 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.4 | 1.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.4 | 2.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.4 | 3.9 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.4 | 1.5 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.4 | 2.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.4 | 3.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.4 | 2.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.4 | 1.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.3 | 2.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 3.9 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.3 | 2.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.3 | 2.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 1.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.0 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 2.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.3 | 0.9 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.3 | 0.9 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 6.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.3 | 4.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 1.9 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 3.6 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 0.7 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 2.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 2.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 2.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 0.7 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 1.5 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.2 | 1.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 2.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 2.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 2.5 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.2 | 2.0 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.2 | 1.8 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 2.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 10.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 2.9 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.2 | 3.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.7 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.6 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 1.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 2.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.5 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.7 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.5 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 3.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 1.4 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 8.1 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.1 | 4.1 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 3.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 2.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 3.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 5.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 2.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.2 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 1.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.3 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 4.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 1.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.5 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 7.6 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 2.8 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.9 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.7 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 1.8 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 8.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 2.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 3.3 | GO:0051225 | spindle assembly(GO:0051225) definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.5 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.1 | 0.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.6 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.0 | 1.8 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 1.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.4 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 1.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 3.1 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 1.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0070073 | regulation of melanocyte differentiation(GO:0045634) clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 1.8 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.7 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 1.2 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 2.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 2.0 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 2.4 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 1.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 2.0 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 7.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.3 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.4 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 5.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:1904861 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) excitatory synapse assembly(GO:1904861) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.4 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 1.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.5 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.1 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.6 | GO:0010564 | regulation of cell cycle process(GO:0010564) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 34.4 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0009615 | response to virus(GO:0009615) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 2.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 2.1 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.6 | 2.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 1.9 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.5 | 1.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 1.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 1.8 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.4 | 2.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.4 | 2.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 3.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 3.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 1.7 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 3.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 2.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.3 | 1.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 6.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 3.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 2.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 1.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 1.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 2.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 4.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 3.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 3.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 1.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 7.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 2.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 7.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.8 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 2.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 2.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 2.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 8.9 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 1.2 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 8.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 8.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 141.8 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.9 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) actomyosin(GO:0042641) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.7 | 2.9 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 5.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 3.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.5 | 1.6 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.5 | 5.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 2.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.4 | 2.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.4 | 1.7 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.4 | 2.0 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.4 | 4.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 2.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.4 | 1.8 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.4 | 2.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 1.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.9 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.3 | 6.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 3.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 6.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 1.7 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 3.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 4.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.9 | GO:0070717 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.2 | 1.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 3.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 1.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 2.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 3.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 0.7 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.2 | 0.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 4.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 3.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 5.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 4.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 5.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 5.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 3.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 2.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.4 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 4.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 5.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 3.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 22.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.1 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 11.3 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 5.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 66.9 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 7.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.6 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 4.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 7.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 2.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 5.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 4.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 6.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 6.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.4 | 3.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 1.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 3.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 5.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 14.7 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 2.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 5.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 2.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 5.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 5.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 3.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.1 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |