Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
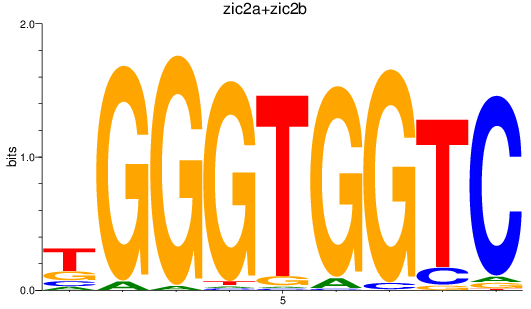
Results for zic2a+zic2b
Z-value: 0.90
Transcription factors associated with zic2a+zic2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic2a
|
ENSDARG00000015554 | zic family member 2 (odd-paired homolog, Drosophila), a |
|
zic2b
|
ENSDARG00000037178 | zic family member 2 (odd-paired homolog, Drosophila) b |
|
zic2b
|
ENSDARG00000112354 | zic family member 2 (odd-paired homolog, Drosophila) b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic2a | dr11_v1_chr9_+_31282161_31282161 | -0.34 | 1.6e-01 | Click! |
| zic2b | dr11_v1_chr1_+_29858032_29858032 | -0.30 | 2.1e-01 | Click! |
Activity profile of zic2a+zic2b motif
Sorted Z-values of zic2a+zic2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_83873 | 6.57 |
ENSDART00000162145
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr20_-_25518488 | 2.95 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr4_-_55785014 | 2.87 |
ENSDART00000181348
|
CT583728.24
|
|
| chr4_-_55801210 | 2.87 |
ENSDART00000186943
|
CT583728.13
|
|
| chr4_-_55829342 | 2.81 |
ENSDART00000185611
|
CT583728.26
|
|
| chr18_-_40684756 | 2.10 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr19_-_10196370 | 2.09 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr5_+_13521081 | 1.97 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr12_+_47162761 | 1.95 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr14_-_49859747 | 1.94 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr13_+_3252950 | 1.90 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr11_-_4235811 | 1.89 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr19_+_9050852 | 1.84 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr12_+_47162456 | 1.54 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr15_-_12229874 | 1.53 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr2_+_51783120 | 1.33 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr2_-_57707039 | 1.32 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr20_-_915234 | 1.29 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr10_+_2876020 | 1.28 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr9_+_54417141 | 1.26 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr21_-_42055872 | 1.25 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr15_-_26844591 | 1.21 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr24_-_26622423 | 1.19 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr12_-_6905375 | 1.18 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr4_-_75175407 | 1.16 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr17_-_34952562 | 1.08 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr9_+_6082793 | 1.06 |
ENSDART00000192045
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr20_-_36671660 | 1.06 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr20_-_7000225 | 1.05 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr19_-_6982272 | 0.99 |
ENSDART00000141913
ENSDART00000150957 |
znf384l
|
zinc finger protein 384 like |
| chr15_-_24178893 | 0.97 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr20_+_1996202 | 0.96 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr22_-_38459316 | 0.96 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr5_-_30151815 | 0.95 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr4_-_2219705 | 0.95 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr1_-_18446161 | 0.94 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr16_-_2558653 | 0.93 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr18_+_9641210 | 0.92 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr3_-_62380146 | 0.91 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr10_+_40235959 | 0.88 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr19_-_22568203 | 0.87 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr23_+_2542158 | 0.83 |
ENSDART00000182197
|
LO017835.1
|
|
| chr24_+_3972260 | 0.78 |
ENSDART00000131753
|
pfkpa
|
phosphofructokinase, platelet a |
| chr3_-_3209432 | 0.78 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr11_-_19097746 | 0.76 |
ENSDART00000103973
|
rhol
|
rhodopsin, like |
| chr11_+_41560792 | 0.76 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr1_-_23370395 | 0.75 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr1_-_58880613 | 0.74 |
ENSDART00000188136
|
CABZ01084501.2
|
|
| chr4_+_10365857 | 0.74 |
ENSDART00000138890
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr16_+_48729947 | 0.74 |
ENSDART00000049051
|
brd2b
|
bromodomain containing 2b |
| chr24_-_21490628 | 0.72 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr14_+_25464681 | 0.70 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr21_+_17110598 | 0.70 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr21_-_45588720 | 0.70 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr10_+_35329751 | 0.69 |
ENSDART00000148043
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr15_+_31276869 | 0.69 |
ENSDART00000156355
|
si:ch211-244e12.7
|
si:ch211-244e12.7 |
| chr18_+_9637744 | 0.68 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr1_-_45042210 | 0.67 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr2_-_33993533 | 0.66 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr11_+_25459697 | 0.65 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr3_-_37351225 | 0.65 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr4_+_4902392 | 0.65 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr7_-_55292116 | 0.65 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr19_-_46566430 | 0.63 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr22_+_9806867 | 0.63 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr21_-_26520629 | 0.63 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr1_+_39597809 | 0.58 |
ENSDART00000122700
|
tenm3
|
teneurin transmembrane protein 3 |
| chr3_-_55537096 | 0.57 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr12_-_124264 | 0.57 |
ENSDART00000181551
|
FO818699.1
|
|
| chr20_+_26095530 | 0.56 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_-_40068350 | 0.56 |
ENSDART00000185292
|
myo15aa
|
myosin XVAa |
| chr22_-_7809904 | 0.56 |
ENSDART00000182269
ENSDART00000097201 ENSDART00000138522 |
si:ch73-44m9.3
si:ch73-44m9.1
|
si:ch73-44m9.3 si:ch73-44m9.1 |
| chr8_-_49304602 | 0.55 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr17_-_7371564 | 0.54 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr17_+_20605013 | 0.52 |
ENSDART00000156878
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr9_+_24159725 | 0.52 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr8_+_29962635 | 0.52 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr4_-_46948863 | 0.51 |
ENSDART00000168835
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_-_30118856 | 0.51 |
ENSDART00000109953
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr21_-_44731865 | 0.51 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr5_+_36439405 | 0.50 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr17_-_8562586 | 0.49 |
ENSDART00000154257
ENSDART00000157308 |
fzd3b
|
frizzled class receptor 3b |
| chr18_-_1143996 | 0.48 |
ENSDART00000189186
ENSDART00000136140 |
hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr17_+_8542203 | 0.48 |
ENSDART00000158873
|
CU462878.2
|
|
| chr8_-_18316482 | 0.48 |
ENSDART00000190156
|
rnf220b
|
ring finger protein 220b |
| chr6_-_49078263 | 0.48 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr13_+_3954540 | 0.47 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr24_-_1356668 | 0.47 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
| chr21_-_45073764 | 0.47 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr6_+_60055168 | 0.46 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr13_+_36958086 | 0.46 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr23_-_27152866 | 0.46 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr21_-_5856050 | 0.46 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr18_-_3527988 | 0.46 |
ENSDART00000157669
|
capn5a
|
calpain 5a |
| chr22_-_20814450 | 0.46 |
ENSDART00000089076
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr11_+_14147913 | 0.45 |
ENSDART00000022823
ENSDART00000154329 |
plppr3b
|
phospholipid phosphatase related 3b |
| chr4_+_38937575 | 0.45 |
ENSDART00000150385
|
znf997
|
zinc finger protein 997 |
| chr18_+_48576527 | 0.44 |
ENSDART00000167555
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr3_+_33745014 | 0.44 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr19_-_28360033 | 0.43 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr11_+_3006124 | 0.42 |
ENSDART00000126071
|
cpne5b
|
copine Vb |
| chr17_-_1407593 | 0.42 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr10_-_13396720 | 0.41 |
ENSDART00000030976
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_3530769 | 0.41 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr4_-_31700186 | 0.41 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr2_-_10703621 | 0.41 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr2_-_12242695 | 0.40 |
ENSDART00000158175
|
gpr158b
|
G protein-coupled receptor 158b |
| chr23_+_30967686 | 0.39 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr9_+_14023386 | 0.39 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr14_-_29859067 | 0.39 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr7_+_17716601 | 0.38 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr2_+_3516913 | 0.37 |
ENSDART00000109346
|
CU693445.1
|
|
| chr5_-_23855447 | 0.37 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr5_-_58112032 | 0.36 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr15_+_26940569 | 0.35 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr17_+_5846202 | 0.35 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr11_+_40657612 | 0.34 |
ENSDART00000183271
|
slc45a1
|
solute carrier family 45, member 1 |
| chr18_+_6558146 | 0.34 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr11_-_23025949 | 0.34 |
ENSDART00000184859
ENSDART00000167818 ENSDART00000046122 ENSDART00000193120 |
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr23_-_11128601 | 0.33 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr5_-_2672492 | 0.33 |
ENSDART00000192337
|
CABZ01072548.1
|
|
| chr17_+_30587333 | 0.32 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr22_-_14128716 | 0.32 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr22_+_26600834 | 0.32 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr5_-_14564878 | 0.31 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr3_-_19091024 | 0.31 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr9_+_21407987 | 0.30 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr16_-_31644545 | 0.30 |
ENSDART00000181634
|
CR855311.5
|
|
| chr7_+_4384863 | 0.29 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr24_-_6078222 | 0.29 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr17_-_7496341 | 0.29 |
ENSDART00000186907
|
si:ch211-278p9.3
|
si:ch211-278p9.3 |
| chr9_-_20372977 | 0.28 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr8_-_18042089 | 0.28 |
ENSDART00000132596
|
dio1
|
iodothyronine deiodinase 1 |
| chr7_-_66864756 | 0.27 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr4_+_10366532 | 0.27 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr7_-_45076131 | 0.27 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr14_-_10371638 | 0.27 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr24_+_24086249 | 0.27 |
ENSDART00000002953
|
lipib
|
lipase, member Ib |
| chr22_-_37611681 | 0.27 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr14_-_40389699 | 0.25 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr6_+_10094061 | 0.25 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr16_-_16481046 | 0.25 |
ENSDART00000158704
|
nbeal2
|
neurobeachin-like 2 |
| chr17_+_34215886 | 0.24 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr24_-_8777781 | 0.24 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
| chr14_+_29609245 | 0.24 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr5_+_7393346 | 0.24 |
ENSDART00000164238
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr3_+_56574623 | 0.23 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr4_+_70967639 | 0.23 |
ENSDART00000159230
|
si:dkeyp-80d11.12
|
si:dkeyp-80d11.12 |
| chr11_-_41130239 | 0.23 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr20_-_28642061 | 0.22 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr23_+_31000243 | 0.21 |
ENSDART00000085263
|
selenoi
|
selenoprotein I |
| chr18_+_6558338 | 0.21 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr11_-_45216684 | 0.20 |
ENSDART00000168621
|
tmc8
|
transmembrane channel-like 8 |
| chr8_+_45294767 | 0.20 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr23_-_14057191 | 0.20 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr8_-_7078575 | 0.19 |
ENSDART00000137413
|
si:ch73-233m11.2
|
si:ch73-233m11.2 |
| chr8_+_26226044 | 0.19 |
ENSDART00000186503
|
wdr6
|
WD repeat domain 6 |
| chr23_+_24989387 | 0.19 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr20_+_27194833 | 0.18 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr17_-_20558961 | 0.17 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr23_+_34005792 | 0.17 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr14_-_48961056 | 0.17 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr25_-_35106751 | 0.16 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr6_+_40922572 | 0.16 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_3372259 | 0.16 |
ENSDART00000140482
|
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr18_+_22994427 | 0.15 |
ENSDART00000173131
ENSDART00000172995 ENSDART00000172923 ENSDART00000141521 ENSDART00000173201 ENSDART00000059961 |
cbfb
|
core-binding factor, beta subunit |
| chr11_+_30162407 | 0.15 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr5_+_20693724 | 0.14 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr21_-_38582061 | 0.14 |
ENSDART00000193891
ENSDART00000124349 |
pof1b
|
POF1B, actin binding protein |
| chr8_-_47800754 | 0.14 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr11_+_14287427 | 0.14 |
ENSDART00000180903
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr25_+_19955598 | 0.13 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr23_-_27015160 | 0.13 |
ENSDART00000189028
|
CU862020.1
|
|
| chr14_+_25465346 | 0.12 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr21_-_22951604 | 0.12 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr23_+_31979602 | 0.12 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_+_25783801 | 0.11 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr13_+_33689740 | 0.10 |
ENSDART00000161904
|
ephx5
|
epoxide hydrolase 5 |
| chr22_+_31207799 | 0.10 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chrM_+_8894 | 0.10 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr20_+_18994465 | 0.10 |
ENSDART00000152778
|
tdh
|
L-threonine dehydrogenase |
| chr20_-_25669813 | 0.09 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr17_+_53294228 | 0.09 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr7_+_3158104 | 0.09 |
ENSDART00000058348
|
CR855393.1
|
|
| chr13_-_38039871 | 0.09 |
ENSDART00000140645
|
CR456624.1
|
|
| chr5_+_19320554 | 0.09 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr3_+_2669813 | 0.09 |
ENSDART00000014205
|
CR388047.1
|
|
| chr17_-_7473890 | 0.08 |
ENSDART00000192910
|
si:ch211-278p9.2
|
si:ch211-278p9.2 |
| chr21_-_21580197 | 0.08 |
ENSDART00000143381
|
or133-3
|
odorant receptor, family H, subfamily 133, member 3 |
| chr12_+_17933775 | 0.08 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr15_+_26941063 | 0.08 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chrM_+_9052 | 0.08 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr2_+_26656283 | 0.07 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr10_+_39248911 | 0.06 |
ENSDART00000170079
ENSDART00000167974 |
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr9_-_3519717 | 0.06 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr4_-_36792144 | 0.06 |
ENSDART00000159147
|
si:dkeyp-87d1.1
|
si:dkeyp-87d1.1 |
| chr8_+_35356944 | 0.06 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_-_45060241 | 0.06 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr17_-_5769196 | 0.05 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr16_+_54263921 | 0.05 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of zic2a+zic2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 1.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 1.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 1.0 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 0.6 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.3 | GO:0042311 | vasodilation(GO:0042311) |
| 0.2 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.6 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 1.0 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.5 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 1.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.8 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.5 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 2.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 3.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.0 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 1.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 1.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 0.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 3.1 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.3 | 1.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 1.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 1.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.5 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 2.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.5 | GO:0048018 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.9 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |