Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
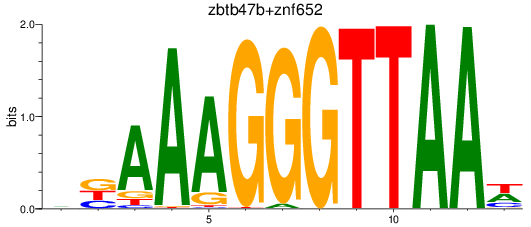
Results for zbtb47b+znf652
Z-value: 1.83
Transcription factors associated with zbtb47b+znf652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf652
|
ENSDARG00000062302 | zinc finger protein 652 |
|
zbtb47b
|
ENSDARG00000079547 | zinc finger and BTB domain containing 47b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf652 | dr11_v1_chr3_+_15773991_15773991 | -0.55 | 1.4e-02 | Click! |
| zbtb47b | dr11_v1_chr24_-_20641000_20641000 | -0.49 | 3.3e-02 | Click! |
Activity profile of zbtb47b+znf652 motif
Sorted Z-values of zbtb47b+znf652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16764751 | 10.26 |
ENSDART00000113862
|
zgc:174154
|
zgc:174154 |
| chr19_-_32928470 | 4.70 |
ENSDART00000141404
ENSDART00000050750 |
rrm2b
|
ribonucleotide reductase M2 b |
| chr9_-_46415847 | 4.70 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr7_+_29167744 | 4.68 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr3_-_54544612 | 3.87 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr22_+_20135443 | 3.76 |
ENSDART00000143641
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr24_+_21514283 | 3.68 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr13_+_7442023 | 3.64 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr20_-_29498178 | 3.48 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr16_-_7228276 | 3.48 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr13_-_33822550 | 3.48 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr7_+_19552381 | 3.25 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_+_38271392 | 3.24 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr7_-_26436436 | 3.20 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr5_+_37978501 | 3.15 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr22_+_18349794 | 3.15 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr4_-_57530173 | 3.08 |
ENSDART00000169785
|
zgc:173702
|
zgc:173702 |
| chr3_-_55650771 | 3.08 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr8_-_18613948 | 3.07 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr13_+_13930263 | 3.02 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr3_-_16411244 | 2.95 |
ENSDART00000140067
|
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr19_+_15444210 | 2.92 |
ENSDART00000142509
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_-_35450661 | 2.90 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr8_-_39822917 | 2.86 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr9_+_15837398 | 2.69 |
ENSDART00000141063
|
si:dkey-103d23.5
|
si:dkey-103d23.5 |
| chr4_-_57001458 | 2.68 |
ENSDART00000158660
|
si:ch211-161m3.1
|
si:ch211-161m3.1 |
| chr19_-_35450857 | 2.67 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr21_+_22892836 | 2.67 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr4_-_44673017 | 2.63 |
ENSDART00000156670
|
si:dkey-7j22.4
|
si:dkey-7j22.4 |
| chr4_-_43280244 | 2.63 |
ENSDART00000150762
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr20_-_27225064 | 2.61 |
ENSDART00000153121
|
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr25_-_35956344 | 2.60 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr4_+_45504471 | 2.58 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr4_-_50544015 | 2.56 |
ENSDART00000155155
|
zgc:174704
|
zgc:174704 |
| chr3_-_1938588 | 2.56 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr16_-_5115993 | 2.52 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr5_-_24517768 | 2.50 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr3_-_29968015 | 2.43 |
ENSDART00000077119
ENSDART00000139310 |
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr4_-_49952636 | 2.38 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr18_-_6862738 | 2.37 |
ENSDART00000192592
|
CT009745.1
|
|
| chr18_-_8380090 | 2.35 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr17_-_35881841 | 2.35 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr4_-_57530817 | 2.33 |
ENSDART00000158435
|
zgc:173702
|
zgc:173702 |
| chr4_-_42294516 | 2.32 |
ENSDART00000133558
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr8_+_2487883 | 2.32 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr22_+_21618121 | 2.31 |
ENSDART00000133939
|
tle2a
|
transducin like enhancer of split 2a |
| chr4_-_69615167 | 2.30 |
ENSDART00000171108
|
si:ch211-120c15.3
|
si:ch211-120c15.3 |
| chr14_-_33308138 | 2.30 |
ENSDART00000136442
ENSDART00000139615 |
sept6
|
septin 6 |
| chr4_-_32180155 | 2.28 |
ENSDART00000164151
|
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr17_-_45733401 | 2.27 |
ENSDART00000185727
|
arf6b
|
ADP-ribosylation factor 6b |
| chr20_+_47143900 | 2.26 |
ENSDART00000153360
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr5_+_69785990 | 2.25 |
ENSDART00000162057
ENSDART00000166893 |
kmt5ab
|
lysine methyltransferase 5Ab |
| chr4_+_33461796 | 2.24 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr4_+_49073583 | 2.23 |
ENSDART00000170022
|
zgc:173705
|
zgc:173705 |
| chr14_-_41369629 | 2.22 |
ENSDART00000173040
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr16_+_23913943 | 2.22 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr4_+_59711338 | 2.19 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr4_-_45301719 | 2.16 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr20_-_30035326 | 2.16 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr9_-_42730672 | 2.13 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr3_+_52953489 | 2.12 |
ENSDART00000125136
|
dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr23_+_7692042 | 2.10 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr18_+_30998472 | 2.10 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr1_+_26110985 | 2.09 |
ENSDART00000054208
|
mtap
|
methylthioadenosine phosphorylase |
| chr23_-_17101360 | 2.07 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr21_+_30721733 | 2.07 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr20_-_32148901 | 2.06 |
ENSDART00000153405
ENSDART00000048537 ENSDART00000152984 |
cep57l1
|
centrosomal protein 57, like 1 |
| chr17_-_53353653 | 2.05 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr4_+_37710744 | 2.05 |
ENSDART00000192791
|
si:dkey-3p4.8
|
si:dkey-3p4.8 |
| chr10_-_21545091 | 2.02 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr4_+_64577406 | 2.02 |
ENSDART00000159754
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_-_36143927 | 2.02 |
ENSDART00000145826
ENSDART00000186186 ENSDART00000164225 |
znf992
|
zinc finger protein 992 |
| chr4_+_58057168 | 2.01 |
ENSDART00000161097
|
si:dkey-57k17.1
|
si:dkey-57k17.1 |
| chr20_+_700616 | 1.98 |
ENSDART00000168166
|
senp6a
|
SUMO1/sentrin specific peptidase 6a |
| chr4_-_70488123 | 1.97 |
ENSDART00000169266
|
si:dkeyp-44b5.5
|
si:dkeyp-44b5.5 |
| chr2_+_26179096 | 1.97 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr24_+_14527935 | 1.96 |
ENSDART00000134846
|
si:dkeyp-73g8.5
|
si:dkeyp-73g8.5 |
| chr24_-_15159658 | 1.95 |
ENSDART00000142473
|
rttn
|
rotatin |
| chr4_+_60934218 | 1.94 |
ENSDART00000150675
|
si:dkey-82i20.2
|
si:dkey-82i20.2 |
| chr15_-_7337537 | 1.94 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr8_+_17168114 | 1.94 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr4_-_52621665 | 1.94 |
ENSDART00000137064
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_-_16545085 | 1.92 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr3_+_24207243 | 1.92 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr3_+_32651697 | 1.92 |
ENSDART00000055338
|
thoc6
|
THO complex 6 |
| chr16_+_5901835 | 1.91 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr9_+_38399912 | 1.89 |
ENSDART00000022246
ENSDART00000145892 |
bcs1l
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr4_-_65302793 | 1.89 |
ENSDART00000158033
|
znf1095
|
zinc finger protein 1095 |
| chr6_+_21684296 | 1.87 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr19_+_43579786 | 1.87 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr4_-_45301250 | 1.87 |
ENSDART00000181753
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr11_+_3959495 | 1.86 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr4_+_58668661 | 1.83 |
ENSDART00000188815
|
CR388148.2
|
|
| chr21_+_21263988 | 1.81 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr4_-_50759477 | 1.80 |
ENSDART00000150246
|
si:ch211-245n8.4
|
si:ch211-245n8.4 |
| chr4_+_18963822 | 1.80 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr4_-_56068511 | 1.79 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr3_-_50865079 | 1.79 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr13_+_47007075 | 1.79 |
ENSDART00000109247
ENSDART00000183205 ENSDART00000180924 ENSDART00000133146 |
anapc1
|
anaphase promoting complex subunit 1 |
| chr22_-_5682494 | 1.78 |
ENSDART00000012686
|
dnase1l4.1
|
deoxyribonuclease 1 like 4, tandem duplicate 1 |
| chr4_+_63818718 | 1.78 |
ENSDART00000161177
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr12_-_33770299 | 1.77 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr4_-_52621232 | 1.76 |
ENSDART00000124451
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_+_54798291 | 1.76 |
ENSDART00000165113
ENSDART00000109624 |
si:dkeyp-82b4.6
|
si:dkeyp-82b4.6 |
| chr24_+_9696760 | 1.76 |
ENSDART00000140200
ENSDART00000187411 |
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr4_-_29833326 | 1.74 |
ENSDART00000150596
|
si:ch211-231i17.4
|
si:ch211-231i17.4 |
| chr23_-_31967554 | 1.74 |
ENSDART00000183973
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr22_-_24946883 | 1.71 |
ENSDART00000193725
ENSDART00000187812 |
si:dkey-4c23.5
|
si:dkey-4c23.5 |
| chr11_+_30744266 | 1.71 |
ENSDART00000132491
ENSDART00000103273 |
wdr83os
|
WD repeat domain 83 opposite strand |
| chr1_-_8553165 | 1.71 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr20_-_20930926 | 1.69 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_-_43279775 | 1.69 |
ENSDART00000183160
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr20_+_28803642 | 1.68 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr15_-_16323750 | 1.67 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr2_+_212059 | 1.66 |
ENSDART00000113021
|
dhx30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr4_+_69191065 | 1.66 |
ENSDART00000170595
|
znf1075
|
zinc finger protein 1075 |
| chr16_-_38609146 | 1.65 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr2_+_29976419 | 1.65 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr4_+_64147241 | 1.64 |
ENSDART00000163509
|
znf1089
|
zinc finger protein 1089 |
| chr4_-_69916852 | 1.64 |
ENSDART00000157665
|
znf1076
|
zinc finger protein 1076 |
| chr9_-_27719998 | 1.63 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr4_-_36144500 | 1.62 |
ENSDART00000170896
|
znf992
|
zinc finger protein 992 |
| chr18_+_20567542 | 1.62 |
ENSDART00000182585
|
bida
|
BH3 interacting domain death agonist |
| chr4_-_50722304 | 1.61 |
ENSDART00000150480
|
znf1110
|
zinc finger protein 1110 |
| chr4_+_58576146 | 1.61 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr1_+_49955869 | 1.59 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr10_-_1718395 | 1.58 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr12_-_4220713 | 1.58 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr23_+_20931030 | 1.58 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr20_+_51061695 | 1.57 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr4_-_64482414 | 1.57 |
ENSDART00000159000
|
znf1105
|
zinc finger protein 1105 |
| chr4_+_58016732 | 1.56 |
ENSDART00000165777
|
CT027609.1
|
|
| chr4_+_63818212 | 1.56 |
ENSDART00000164929
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr25_-_14424406 | 1.56 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr4_+_62262253 | 1.55 |
ENSDART00000166022
|
si:dkeyp-35e5.10
|
si:dkeyp-35e5.10 |
| chr18_-_44623675 | 1.55 |
ENSDART00000005431
|
psmd8
|
proteasome 26S subunit, non-ATPase 8 |
| chr4_+_45504938 | 1.55 |
ENSDART00000145958
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr7_-_16034324 | 1.54 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr5_+_58455488 | 1.54 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr18_-_17075098 | 1.54 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr20_+_25625872 | 1.54 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr20_-_20355577 | 1.53 |
ENSDART00000018500
|
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr4_+_64577910 | 1.53 |
ENSDART00000128164
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_+_53183373 | 1.53 |
ENSDART00000176617
|
znf1037
|
zinc finger protein 1037 |
| chr10_+_39893439 | 1.51 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr14_+_34966598 | 1.50 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr4_-_69917450 | 1.49 |
ENSDART00000168915
|
znf1076
|
zinc finger protein 1076 |
| chr8_+_15251448 | 1.49 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr23_-_40194732 | 1.48 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr10_-_40791977 | 1.46 |
ENSDART00000109020
|
zgc:113625
|
zgc:113625 |
| chr7_-_52963493 | 1.45 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr11_-_39118882 | 1.45 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr14_+_32852388 | 1.44 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr4_-_31795106 | 1.41 |
ENSDART00000164175
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr6_+_21001264 | 1.40 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr24_-_27423697 | 1.40 |
ENSDART00000132590
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr19_+_26640096 | 1.40 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr3_-_43959873 | 1.40 |
ENSDART00000161582
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr20_-_20931197 | 1.40 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_-_57994891 | 1.39 |
ENSDART00000168657
|
znf1097
|
zinc finger protein 1097 |
| chr4_+_43408004 | 1.39 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr5_-_38643188 | 1.38 |
ENSDART00000144972
|
si:ch211-271e10.1
|
si:ch211-271e10.1 |
| chr4_-_29769304 | 1.38 |
ENSDART00000134990
|
znf1119
|
zinc finger protein 1119 |
| chr5_+_51833305 | 1.38 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr17_-_6536305 | 1.37 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr6_-_2627488 | 1.37 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr16_-_27677930 | 1.36 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr6_-_49173891 | 1.35 |
ENSDART00000132867
|
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr17_+_26815021 | 1.35 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr1_+_513986 | 1.33 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr1_-_31140096 | 1.33 |
ENSDART00000172243
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr13_+_502230 | 1.31 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr4_+_59748607 | 1.29 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr4_+_38170708 | 1.28 |
ENSDART00000168900
|
znf1071
|
zinc finger protein 1071 |
| chr4_+_35901451 | 1.28 |
ENSDART00000131142
|
znf1088
|
zinc finger protein 1088 |
| chr3_-_12970418 | 1.28 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr4_+_28962774 | 1.27 |
ENSDART00000150629
|
si:dkey-23a23.3
|
si:dkey-23a23.3 |
| chr4_+_69559692 | 1.27 |
ENSDART00000164383
|
znf993
|
zinc finger protein 993 |
| chr5_-_43935460 | 1.27 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr13_-_18637244 | 1.27 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr11_-_3535537 | 1.26 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr7_-_26306954 | 1.26 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr1_+_31573225 | 1.26 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr6_+_31368071 | 1.24 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr4_+_29532731 | 1.24 |
ENSDART00000114649
|
znf1140
|
zinc finger protein 1140 |
| chr3_+_50201240 | 1.24 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr4_+_41528590 | 1.23 |
ENSDART00000164004
|
znf1094
|
zinc finger protein 1094 |
| chr2_-_55853943 | 1.23 |
ENSDART00000122576
|
rx2
|
retinal homeobox gene 2 |
| chr4_+_62337473 | 1.22 |
ENSDART00000166513
ENSDART00000160761 |
znf1079
|
zinc finger protein 1079 |
| chr16_-_31661536 | 1.22 |
ENSDART00000169973
|
wu:fd46c06
|
wu:fd46c06 |
| chr13_+_51710725 | 1.22 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr4_+_64102312 | 1.22 |
ENSDART00000157859
|
znf1103
|
zinc finger protein 1103 |
| chr20_-_26537171 | 1.20 |
ENSDART00000061926
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr5_-_69716501 | 1.20 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr7_-_40993456 | 1.19 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr5_-_64431927 | 1.18 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr7_-_67248829 | 1.17 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr14_+_20929586 | 1.17 |
ENSDART00000106198
ENSDART00000166366 |
zgc:66433
|
zgc:66433 |
| chr20_+_52584532 | 1.17 |
ENSDART00000138641
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr4_+_59589201 | 1.17 |
ENSDART00000150576
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb47b+znf652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.8 | 2.5 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.7 | 8.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 3.2 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.6 | 2.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.6 | 3.0 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.5 | 1.6 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.5 | 1.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.5 | 1.6 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.5 | 1.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 | 2.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.5 | 2.9 | GO:0097065 | anterior head development(GO:0097065) |
| 0.4 | 1.3 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.4 | 1.8 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.4 | 1.7 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.4 | 1.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 2.1 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.4 | 2.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 2.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 4.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 1.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 2.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 2.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.9 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.4 | 1.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.4 | 3.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 1.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.4 | 1.8 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.3 | 2.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 3.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 2.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 1.6 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.3 | 1.1 | GO:0021543 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.2 | 2.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.0 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.2 | 3.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 3.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 1.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 3.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 1.9 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 1.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 1.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 1.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.6 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.9 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 1.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.6 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 2.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 2.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.8 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 1.2 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 1.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 1.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 1.7 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 4.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.4 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 2.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 3.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 0.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.5 | GO:0071456 | response to glucose(GO:0009749) cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 2.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 3.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.0 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.1 | 1.0 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.9 | GO:0030316 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) osteoclast differentiation(GO:0030316) |
| 0.0 | 1.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.8 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 1.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 122.9 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.8 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 1.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 3.1 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.0 | 0.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 1.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 2.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.8 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0031100 | organ regeneration(GO:0031100) pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 2.1 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.6 | 2.5 | GO:0031511 | condensed chromosome inner kinetochore(GO:0000939) Mis6-Sim4 complex(GO:0031511) |
| 0.4 | 1.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 1.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 1.7 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.3 | 1.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 3.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 2.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 3.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 4.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 6.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.6 | GO:0031968 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.2 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 88.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.0 | 2.9 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.6 | 3.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 2.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 1.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.5 | 1.5 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 1.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 1.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 1.7 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 4.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 3.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 2.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 2.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 2.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 3.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.7 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 1.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 2.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.5 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 1.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 3.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 137.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 1.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 7.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 3.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.1 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 3.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 2.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 1.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 2.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 5.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 3.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 5.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 4.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 1.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.7 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 1.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |