Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
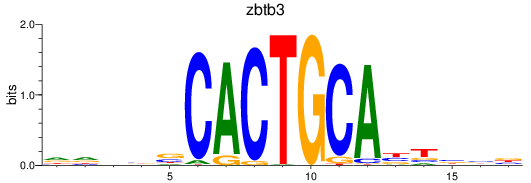
Results for zbtb3
Z-value: 0.55
Transcription factors associated with zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb3
|
ENSDARG00000036235 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb3 | dr11_v1_chr14_+_35405518_35405518 | -0.36 | 1.2e-01 | Click! |
Activity profile of zbtb3 motif
Sorted Z-values of zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_52398205 | 2.93 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr3_-_61185746 | 2.00 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr7_-_34225011 | 1.87 |
ENSDART00000049588
|
crybgx
|
crystallin beta gamma X |
| chr2_+_45643122 | 1.75 |
ENSDART00000138179
|
fndc7rs4
|
fibronectin type III domain containing 7, related sequence 4 |
| chr2_-_27329214 | 1.30 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr16_-_24598042 | 0.98 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr13_-_2215213 | 0.95 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr24_-_26383978 | 0.86 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr7_-_26087807 | 0.83 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr12_+_10116912 | 0.82 |
ENSDART00000189630
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr2_-_985417 | 0.77 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr16_-_42872571 | 0.68 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr2_+_5621529 | 0.67 |
ENSDART00000144187
|
fgf12a
|
fibroblast growth factor 12a |
| chr6_-_38419318 | 0.62 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr15_-_12113045 | 0.62 |
ENSDART00000159879
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr1_+_44814322 | 0.61 |
ENSDART00000059227
|
ndufs8a
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8a |
| chr1_-_22687913 | 0.58 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr3_+_24458899 | 0.57 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr22_-_29336268 | 0.56 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr7_-_36358735 | 0.53 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr8_-_54304381 | 0.52 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr3_+_61185660 | 0.50 |
ENSDART00000167114
|
CR352263.1
|
|
| chr9_+_14023386 | 0.46 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr19_+_25478203 | 0.45 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr16_+_34531486 | 0.43 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr25_-_35542739 | 0.42 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr3_-_24458281 | 0.41 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr6_-_39006449 | 0.41 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr17_+_30450163 | 0.40 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr15_-_576135 | 0.40 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr7_-_6592142 | 0.39 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr23_+_39558508 | 0.39 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr8_+_3820134 | 0.35 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr10_-_4375190 | 0.34 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr16_-_32013913 | 0.33 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr15_+_863931 | 0.33 |
ENSDART00000154083
|
si:dkey-7i4.12
|
si:dkey-7i4.12 |
| chr11_+_1805421 | 0.32 |
ENSDART00000173143
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr6_+_7507390 | 0.32 |
ENSDART00000149285
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr2_+_15776408 | 0.31 |
ENSDART00000176628
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr14_-_32258759 | 0.31 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr7_-_57949117 | 0.29 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr2_+_45484183 | 0.28 |
ENSDART00000183490
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr24_+_12740848 | 0.27 |
ENSDART00000060825
|
si:ch211-196f5.2
|
si:ch211-196f5.2 |
| chr22_-_25469751 | 0.27 |
ENSDART00000171670
|
CR769772.4
|
|
| chr7_-_12789251 | 0.26 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr11_+_10548171 | 0.26 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr3_+_24458204 | 0.25 |
ENSDART00000155028
ENSDART00000153551 |
cbx6b
|
chromobox homolog 6b |
| chr3_+_49397115 | 0.25 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr1_-_44704261 | 0.25 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr22_-_25502977 | 0.22 |
ENSDART00000181749
|
CR769772.4
|
|
| chr1_+_59007536 | 0.21 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr8_+_54135642 | 0.21 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr7_-_48263516 | 0.21 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr17_-_29311835 | 0.20 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr23_+_6232895 | 0.20 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr12_+_48220584 | 0.20 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr1_+_532766 | 0.20 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr7_+_33279618 | 0.20 |
ENSDART00000173706
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr21_-_4923427 | 0.19 |
ENSDART00000181489
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr1_-_633356 | 0.18 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr21_+_45366229 | 0.18 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr10_+_44641599 | 0.18 |
ENSDART00000172128
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr5_+_43458304 | 0.18 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr16_+_13898399 | 0.17 |
ENSDART00000140946
|
si:ch211-149k23.9
|
si:ch211-149k23.9 |
| chr7_+_24522308 | 0.17 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr7_+_34492744 | 0.16 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr11_-_42230491 | 0.16 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr2_+_47718605 | 0.16 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr19_+_40122370 | 0.16 |
ENSDART00000151276
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr14_+_6546516 | 0.14 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr24_-_24146875 | 0.13 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr18_-_43880020 | 0.13 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr13_+_23282549 | 0.12 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr18_+_37015185 | 0.12 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr2_+_31454562 | 0.11 |
ENSDART00000136360
|
TMEM236
|
si:dkey-32m20.1 |
| chr3_-_5353442 | 0.11 |
ENSDART00000188808
|
trim35-7
|
tripartite motif containing 35-7 |
| chr19_+_917852 | 0.11 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr3_-_3019979 | 0.11 |
ENSDART00000111174
|
zgc:172090
|
zgc:172090 |
| chr13_+_771403 | 0.10 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr2_-_54639964 | 0.10 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr19_+_42770041 | 0.10 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr23_-_15878879 | 0.09 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr22_-_11136625 | 0.09 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr20_-_6131686 | 0.09 |
ENSDART00000145964
ENSDART00000086578 ENSDART00000164090 |
pum2
|
pumilio RNA-binding family member 2 |
| chr2_+_15776649 | 0.09 |
ENSDART00000156535
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr15_-_2632891 | 0.09 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr21_-_11791909 | 0.09 |
ENSDART00000180893
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr2_+_68789 | 0.08 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr9_+_307863 | 0.08 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr9_-_18814737 | 0.08 |
ENSDART00000131267
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr7_+_39079071 | 0.08 |
ENSDART00000181845
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr18_+_7611298 | 0.07 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr22_-_5051689 | 0.07 |
ENSDART00000113521
|
matk
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_29005622 | 0.06 |
ENSDART00000170162
|
CR376738.2
|
|
| chr23_-_32404022 | 0.06 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr17_-_868004 | 0.05 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr22_+_3238474 | 0.05 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr17_-_36841981 | 0.05 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr4_-_68913650 | 0.04 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr17_+_52524452 | 0.04 |
ENSDART00000016614
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr6_+_58698475 | 0.03 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr17_-_36842267 | 0.03 |
ENSDART00000042203
ENSDART00000176989 |
myo6b
|
myosin VIb |
| chr3_+_17456428 | 0.03 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr1_+_58079053 | 0.03 |
ENSDART00000141678
|
si:ch211-114l13.3
|
si:ch211-114l13.3 |
| chr8_-_8446668 | 0.03 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr16_-_30969294 | 0.02 |
ENSDART00000147890
|
dgat1b
|
diacylglycerol O-acyltransferase 1b |
| chr14_+_1355857 | 0.02 |
ENSDART00000188008
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr13_+_22964868 | 0.02 |
ENSDART00000142129
|
tacr2
|
tachykinin receptor 2 |
| chr14_-_33858214 | 0.02 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr20_+_27713210 | 0.01 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr7_+_30051488 | 0.01 |
ENSDART00000169410
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr15_-_19705707 | 0.00 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 0.8 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 0.5 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.8 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.5 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |