Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
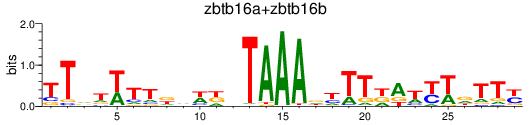
Results for zbtb16a+zbtb16b
Z-value: 0.86
Transcription factors associated with zbtb16a+zbtb16b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb16a
|
ENSDARG00000007184 | zinc finger and BTB domain containing 16a |
|
zbtb16b
|
ENSDARG00000074526 | zinc finger and BTB domain containing 16b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb16b | dr11_v1_chr15_-_18361475_18361475 | 0.39 | 1.0e-01 | Click! |
| zbtb16a | dr11_v1_chr21_-_23307653_23307796 | 0.18 | 4.6e-01 | Click! |
Activity profile of zbtb16a+zbtb16b motif
Sorted Z-values of zbtb16a+zbtb16b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_16853464 | 1.13 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr8_+_39724138 | 1.11 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr1_+_34203817 | 0.98 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr8_-_6825588 | 0.92 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr1_+_44173245 | 0.88 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr12_-_20796430 | 0.86 |
ENSDART00000064339
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr10_-_17745345 | 0.80 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr6_-_53885514 | 0.79 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr24_+_35947077 | 0.79 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr24_-_33578392 | 0.76 |
ENSDART00000079476
|
LO018309.1
|
|
| chr20_-_47051996 | 0.76 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr12_+_2870671 | 0.74 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr2_-_37888429 | 0.74 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr21_+_17541634 | 0.70 |
ENSDART00000191700
ENSDART00000163238 |
stom
|
stomatin |
| chr9_+_1139378 | 0.69 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr18_+_15841449 | 0.68 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr19_-_22387141 | 0.68 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr23_-_9492266 | 0.68 |
ENSDART00000091852
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr9_-_49464993 | 0.64 |
ENSDART00000181471
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr2_-_122154 | 0.63 |
ENSDART00000156248
ENSDART00000004071 |
znfl2a
|
zinc finger-like gene 2a |
| chr8_+_3820134 | 0.63 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr16_-_54078662 | 0.61 |
ENSDART00000100602
|
cdk14
|
cyclin-dependent kinase 14 |
| chr12_+_28854963 | 0.61 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr12_-_20796658 | 0.61 |
ENSDART00000181253
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr18_-_44527124 | 0.60 |
ENSDART00000189471
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr14_+_15620640 | 0.58 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr15_-_25093680 | 0.55 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr5_+_5398966 | 0.54 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr2_-_1548330 | 0.54 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr13_-_37474989 | 0.54 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr11_+_13224281 | 0.54 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr20_+_31270761 | 0.52 |
ENSDART00000176187
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr17_+_16565185 | 0.51 |
ENSDART00000136874
|
foxn3
|
forkhead box N3 |
| chr13_+_20007366 | 0.50 |
ENSDART00000147217
|
atrnl1a
|
attractin-like 1a |
| chr24_-_36238054 | 0.50 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr23_+_40951443 | 0.50 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr9_+_50316921 | 0.50 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr5_+_66250856 | 0.50 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr25_-_998096 | 0.50 |
ENSDART00000164082
|
znf609a
|
zinc finger protein 609a |
| chr15_-_26931541 | 0.49 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr15_-_20190052 | 0.48 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr22_-_29204960 | 0.48 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr4_-_20292821 | 0.48 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr2_-_55298075 | 0.48 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr10_+_44146287 | 0.48 |
ENSDART00000162176
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr17_+_16564921 | 0.46 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr14_-_7045009 | 0.46 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr4_-_76270779 | 0.46 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr15_-_25094026 | 0.45 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr23_+_42810055 | 0.45 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr2_+_37875789 | 0.44 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr21_-_293146 | 0.44 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr8_-_45867358 | 0.44 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr2_+_54696042 | 0.43 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr14_-_24095414 | 0.43 |
ENSDART00000172747
ENSDART00000173146 |
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr6_+_39085969 | 0.42 |
ENSDART00000004240
|
prss60.1
|
protease, serine, 60.1 |
| chr11_+_691734 | 0.41 |
ENSDART00000191463
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr20_-_2949028 | 0.41 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr16_+_27593698 | 0.41 |
ENSDART00000145203
|
si:ch211-197h24.8
|
si:ch211-197h24.8 |
| chr11_-_37359416 | 0.40 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr17_-_35352690 | 0.40 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr20_-_630637 | 0.39 |
ENSDART00000160454
|
si:dkey-121j17.5
|
si:dkey-121j17.5 |
| chr15_+_3219134 | 0.39 |
ENSDART00000113532
|
LO017656.1
|
|
| chr12_+_48841419 | 0.39 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr8_+_1651821 | 0.39 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr2_+_15203322 | 0.38 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr2_+_44512324 | 0.38 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr13_-_5978433 | 0.37 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr9_-_7378566 | 0.37 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr8_-_44004135 | 0.37 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr17_-_23603263 | 0.37 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr20_+_13175379 | 0.37 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr5_+_38726854 | 0.36 |
ENSDART00000138484
ENSDART00000142867 |
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr4_-_17725008 | 0.36 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr21_-_9383974 | 0.36 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr8_+_53563949 | 0.36 |
ENSDART00000190022
ENSDART00000190927 |
CU861886.1
|
|
| chr24_-_33873451 | 0.35 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr10_+_575929 | 0.35 |
ENSDART00000129856
|
smad4a
|
SMAD family member 4a |
| chr22_+_19277361 | 0.35 |
ENSDART00000190075
ENSDART00000141097 ENSDART00000136943 |
si:dkey-21e2.13
|
si:dkey-21e2.13 |
| chr2_+_21048661 | 0.35 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr16_-_2414063 | 0.34 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr23_+_2361184 | 0.34 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr19_-_27588842 | 0.34 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr12_+_48841182 | 0.34 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr5_-_71765780 | 0.34 |
ENSDART00000011955
|
gpsm1b
|
G protein signaling modulator 1b |
| chr6_-_12722360 | 0.33 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr3_+_23248542 | 0.33 |
ENSDART00000185765
ENSDART00000192332 |
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr3_-_1938588 | 0.33 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr19_-_425145 | 0.33 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr4_+_5868034 | 0.33 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr10_-_7666810 | 0.32 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr7_-_24838857 | 0.32 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr25_-_997894 | 0.32 |
ENSDART00000169011
|
znf609a
|
zinc finger protein 609a |
| chr7_+_61184104 | 0.32 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr16_-_29146624 | 0.32 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr19_+_30990129 | 0.31 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr11_+_6902946 | 0.31 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr17_+_39242437 | 0.31 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr13_-_27388097 | 0.31 |
ENSDART00000187472
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr17_+_41343167 | 0.30 |
ENSDART00000156405
|
mocs1
|
molybdenum cofactor synthesis 1 |
| chr20_+_41906960 | 0.30 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr13_+_19283936 | 0.30 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr19_+_43753995 | 0.29 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr9_+_52398531 | 0.29 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr19_+_15440841 | 0.29 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_-_68826177 | 0.28 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr24_-_36175365 | 0.28 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr19_+_15441022 | 0.28 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_+_6902306 | 0.28 |
ENSDART00000075993
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr21_-_2341937 | 0.28 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr20_+_23173710 | 0.28 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr18_-_42071876 | 0.28 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr3_+_62161184 | 0.28 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr2_-_9260002 | 0.27 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr2_-_11027258 | 0.27 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr2_-_23931536 | 0.27 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr13_+_44857087 | 0.26 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr3_-_45848043 | 0.26 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr14_-_21123551 | 0.26 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr13_+_2617555 | 0.26 |
ENSDART00000162208
|
plpp4
|
phospholipid phosphatase 4 |
| chr8_+_28724692 | 0.26 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr14_+_24840669 | 0.25 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr3_-_50136424 | 0.24 |
ENSDART00000188843
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr5_-_24642026 | 0.24 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr22_-_16403602 | 0.24 |
ENSDART00000183417
|
lama3
|
laminin, alpha 3 |
| chr18_-_2639351 | 0.24 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr8_-_50133064 | 0.24 |
ENSDART00000030064
|
antxr1a
|
anthrax toxin receptor 1a |
| chr23_+_19213472 | 0.23 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr25_-_27665978 | 0.23 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr2_-_1443692 | 0.23 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr4_-_69550757 | 0.23 |
ENSDART00000168344
|
znf1101
|
zinc finger protein 1101 |
| chr8_-_16499508 | 0.23 |
ENSDART00000108659
|
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr7_-_24520866 | 0.23 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr4_+_44808133 | 0.22 |
ENSDART00000184430
|
znf1107
|
zinc finger protein 1107 |
| chr25_+_34888886 | 0.22 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr22_+_1568306 | 0.22 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr21_-_35082715 | 0.22 |
ENSDART00000146454
|
adrb2b
|
adrenoceptor beta 2, surface b |
| chr25_-_7709797 | 0.22 |
ENSDART00000157076
|
prdm11
|
PR domain containing 11 |
| chr9_+_32178050 | 0.22 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr8_+_31716872 | 0.22 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr3_-_45848257 | 0.22 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr5_-_18046053 | 0.21 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr8_-_11546175 | 0.21 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr20_-_32752457 | 0.21 |
ENSDART00000153287
|
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr15_-_18124678 | 0.21 |
ENSDART00000062523
|
cryaba
|
crystallin, alpha B, a |
| chr4_-_44673017 | 0.20 |
ENSDART00000156670
|
si:dkey-7j22.4
|
si:dkey-7j22.4 |
| chr8_+_19624589 | 0.20 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr4_-_59836867 | 0.20 |
ENSDART00000142868
|
CR846087.3
|
|
| chr4_-_1839192 | 0.20 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr2_-_9259283 | 0.19 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr4_-_75997108 | 0.19 |
ENSDART00000163113
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr4_+_37936249 | 0.19 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr9_+_9441453 | 0.19 |
ENSDART00000081859
ENSDART00000188567 ENSDART00000143101 |
maats1
|
MYCBP-associated, testis expressed 1 |
| chr7_-_72426484 | 0.19 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr4_+_49506359 | 0.19 |
ENSDART00000154478
|
si:dkey-5i16.5
|
si:dkey-5i16.5 |
| chr4_-_38421217 | 0.18 |
ENSDART00000164517
|
znf1092
|
szinc finger protein 1092 |
| chr15_+_31820536 | 0.18 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr15_-_17960228 | 0.18 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr9_+_34232503 | 0.18 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr5_+_13245589 | 0.18 |
ENSDART00000193618
|
CR933528.1
|
|
| chr22_+_1170294 | 0.18 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr10_-_32494304 | 0.18 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr21_+_45510448 | 0.18 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr8_+_53565471 | 0.18 |
ENSDART00000179055
|
CU861886.1
|
|
| chr8_+_20679759 | 0.18 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr16_+_4838808 | 0.18 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr21_-_32436679 | 0.17 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr12_-_45197038 | 0.17 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr9_+_42095220 | 0.17 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr18_+_30508729 | 0.17 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr2_-_7757273 | 0.17 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr6_+_55819038 | 0.17 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr11_+_41459408 | 0.17 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr6_-_14040136 | 0.17 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr24_-_2450597 | 0.16 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr8_-_32354677 | 0.16 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr4_-_37587110 | 0.16 |
ENSDART00000169302
|
znf1100
|
zinc finger protein 1100 |
| chr22_-_29205327 | 0.16 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr6_+_6828167 | 0.16 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr11_+_26403334 | 0.16 |
ENSDART00000171781
|
fer1l4
|
fer-1-like family member 4 |
| chr10_-_32494499 | 0.16 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr21_-_13051613 | 0.15 |
ENSDART00000190777
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr8_+_10350122 | 0.15 |
ENSDART00000186650
ENSDART00000098009 |
tbc1d22b
|
TBC1 domain family, member 22B |
| chr18_-_16885362 | 0.15 |
ENSDART00000132778
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr10_+_7636811 | 0.15 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr17_-_24714837 | 0.15 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr2_+_5841108 | 0.15 |
ENSDART00000136180
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr6_-_49510553 | 0.15 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr22_+_6740039 | 0.15 |
ENSDART00000144122
|
CT583625.2
|
|
| chr7_-_12789251 | 0.14 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr5_+_58421536 | 0.14 |
ENSDART00000173410
|
ccdc15
|
coiled-coil domain containing 15 |
| chr21_+_10712823 | 0.14 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr20_+_36628059 | 0.14 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr13_+_40686133 | 0.13 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr8_-_50132860 | 0.13 |
ENSDART00000149964
|
antxr1a
|
anthrax toxin receptor 1a |
| chr9_+_45227028 | 0.13 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr22_-_27113332 | 0.13 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr19_+_4059200 | 0.13 |
ENSDART00000161676
ENSDART00000172424 ENSDART00000161804 |
btr25
|
bloodthirsty-related gene family, member 25 |
| chr4_-_1839352 | 0.13 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr16_-_45209684 | 0.12 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb16a+zbtb16b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.4 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.5 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.2 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.5 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.9 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.2 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.9 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.4 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.4 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.5 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.8 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.7 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0035108 | hypothalamus cell differentiation(GO:0021979) limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.5 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.3 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.2 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.4 | GO:0008191 | protease binding(GO:0002020) metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |