Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
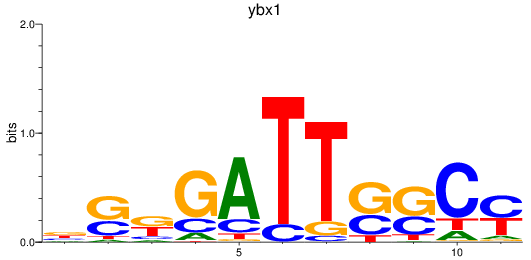
Results for ybx1
Z-value: 0.57
Transcription factors associated with ybx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ybx1
|
ENSDARG00000004757 | Y box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ybx1 | dr11_v1_chr8_-_47152001_47152086 | -0.54 | 1.8e-02 | Click! |
Activity profile of ybx1 motif
Sorted Z-values of ybx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_44883805 | 7.51 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr2_-_42091926 | 2.78 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr20_-_52939501 | 2.26 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr17_+_45305645 | 1.48 |
ENSDART00000172488
|
capn3a
|
calpain 3a, (p94) |
| chr25_-_4148719 | 1.24 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr1_-_20928772 | 1.22 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr12_+_46960579 | 1.22 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr3_+_1167026 | 1.18 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr6_-_35446110 | 1.15 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr18_+_50089000 | 1.13 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr25_-_19433244 | 1.06 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr6_-_48094342 | 1.03 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr6_+_53349966 | 1.00 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr23_-_41651759 | 0.98 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr11_-_44543082 | 0.97 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr18_+_907266 | 0.94 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr6_-_52156427 | 0.94 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr7_-_6466119 | 0.91 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr20_-_7176809 | 0.89 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr10_+_44903676 | 0.89 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr21_-_5007109 | 0.89 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr2_-_4797512 | 0.89 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr3_+_3810919 | 0.89 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr19_-_657439 | 0.88 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr24_-_39772045 | 0.88 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr4_-_240737 | 0.86 |
ENSDART00000166186
|
RERG
|
si:cabz01085950.1 |
| chr24_+_7631797 | 0.86 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr8_+_54081819 | 0.85 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr18_+_660578 | 0.83 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr13_-_856525 | 0.83 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr24_+_24923166 | 0.81 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr3_-_13147310 | 0.78 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr5_+_5398966 | 0.76 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr23_+_2778813 | 0.76 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr9_+_27411502 | 0.75 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr19_-_6134802 | 0.75 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_+_38529263 | 0.75 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr21_-_26406244 | 0.74 |
ENSDART00000137312
ENSDART00000077200 |
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr10_+_6010570 | 0.74 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr12_+_1469090 | 0.74 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr23_-_35483163 | 0.73 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr23_+_19564392 | 0.73 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr20_-_53366137 | 0.71 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr13_-_856701 | 0.71 |
ENSDART00000140423
|
TMEM14A
|
zgc:163080 |
| chr10_+_22782522 | 0.69 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr4_+_11375894 | 0.69 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr6_+_37752781 | 0.67 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr9_-_30145080 | 0.66 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr15_-_3094516 | 0.66 |
ENSDART00000179719
|
slitrk5a
|
SLIT and NTRK-like family, member 5a |
| chr5_+_20030414 | 0.65 |
ENSDART00000181430
ENSDART00000047841 ENSDART00000182813 |
sgsm1a
|
small G protein signaling modulator 1a |
| chr3_-_61218272 | 0.64 |
ENSDART00000133091
ENSDART00000055068 |
pvalb5
|
parvalbumin 5 |
| chr24_-_41220538 | 0.63 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr13_+_22264914 | 0.62 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr25_+_4541211 | 0.62 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr21_+_3897680 | 0.61 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr24_-_24271629 | 0.61 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr7_+_1521834 | 0.61 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr10_-_35236949 | 0.60 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr4_-_28158335 | 0.59 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr19_+_42806812 | 0.58 |
ENSDART00000108775
ENSDART00000151653 |
ubp1
|
upstream binding protein 1 |
| chr21_-_293146 | 0.57 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr15_+_1148074 | 0.57 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr14_+_3038473 | 0.56 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr8_-_1219815 | 0.56 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr13_-_3370638 | 0.54 |
ENSDART00000029649
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr21_+_5888641 | 0.53 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr16_+_68069 | 0.53 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr7_+_5906327 | 0.52 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr3_+_36541003 | 0.52 |
ENSDART00000164514
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr15_+_43166511 | 0.51 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr16_+_45930962 | 0.50 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr18_+_27515640 | 0.50 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr15_-_19250543 | 0.48 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr25_+_10458990 | 0.48 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr25_-_35542739 | 0.47 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr17_-_43286903 | 0.47 |
ENSDART00000176637
|
EML5
|
si:dkey-1f12.3 |
| chr22_+_17205608 | 0.47 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr12_+_40905427 | 0.47 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr9_-_2572790 | 0.46 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr4_-_76370630 | 0.46 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr7_-_6364168 | 0.46 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr11_-_42554290 | 0.46 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr8_-_44015210 | 0.45 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr9_+_45227028 | 0.44 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr11_+_77526 | 0.44 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr18_-_26894732 | 0.44 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr12_+_1469327 | 0.44 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr25_-_35150933 | 0.44 |
ENSDART00000129254
|
HIST2H3C
|
zgc:173552 |
| chr13_-_36050303 | 0.43 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr2_-_58193381 | 0.43 |
ENSDART00000159040
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr17_-_6514962 | 0.43 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr15_-_47336811 | 0.43 |
ENSDART00000182730
|
anapc15
|
anaphase promoting complex subunit 15 |
| chr7_-_69636502 | 0.42 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr10_-_15644904 | 0.42 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr16_+_46695777 | 0.42 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr24_-_21404367 | 0.41 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr23_+_14590767 | 0.41 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr19_-_1855368 | 0.40 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr9_+_22677503 | 0.40 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr7_-_73834812 | 0.40 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr14_-_12071447 | 0.38 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr19_-_30562648 | 0.37 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr9_-_2573121 | 0.37 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr23_+_14590483 | 0.37 |
ENSDART00000088359
ENSDART00000184868 |
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr20_-_40487208 | 0.36 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr2_+_24867534 | 0.36 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr2_+_47708853 | 0.36 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr3_-_58543658 | 0.35 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr6_-_218882 | 0.34 |
ENSDART00000008398
|
c1qtnf6b
|
C1q and TNF related 6b |
| chr22_-_11493236 | 0.34 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr16_+_4838808 | 0.34 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_-_10751491 | 0.34 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr20_+_16743056 | 0.34 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr23_+_6795531 | 0.34 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr1_+_6817292 | 0.34 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr22_+_38173960 | 0.33 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr21_+_944715 | 0.32 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr14_-_40821411 | 0.32 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr5_+_45138934 | 0.31 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_2499627 | 0.30 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr25_-_35110695 | 0.30 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr21_+_244503 | 0.29 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr18_-_49116382 | 0.29 |
ENSDART00000174157
|
BX663503.3
|
|
| chr8_+_36288261 | 0.29 |
ENSDART00000191281
|
CU929676.1
|
|
| chr10_-_7858553 | 0.29 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr19_-_43004356 | 0.28 |
ENSDART00000098101
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr22_+_30543437 | 0.28 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr2_+_37480669 | 0.28 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr7_-_13884610 | 0.27 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr24_+_7708652 | 0.27 |
ENSDART00000066816
|
btr33
|
bloodthirsty-related gene family, member 33 |
| chr3_-_8765165 | 0.27 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr6_+_3710865 | 0.26 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr20_-_49889111 | 0.26 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr23_+_45579497 | 0.26 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr13_+_2394534 | 0.26 |
ENSDART00000172535
ENSDART00000006990 |
elovl5
|
ELOVL fatty acid elongase 5 |
| chr15_+_25439106 | 0.25 |
ENSDART00000156252
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr5_-_16983336 | 0.24 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr18_-_2549198 | 0.24 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr6_+_52350443 | 0.24 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr19_+_770300 | 0.24 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr1_+_25801648 | 0.24 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr5_+_45139196 | 0.24 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr20_-_3403033 | 0.23 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr15_+_25438714 | 0.23 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr2_+_24868010 | 0.23 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr7_-_6431158 | 0.22 |
ENSDART00000173199
|
si:ch1073-153i20.5
|
si:ch1073-153i20.5 |
| chr1_+_1599979 | 0.22 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr7_+_16033923 | 0.21 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr3_+_62353650 | 0.21 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr24_+_34606966 | 0.21 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr18_+_2563756 | 0.21 |
ENSDART00000164147
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr22_-_4407871 | 0.21 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr3_+_59051503 | 0.21 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr16_-_44349845 | 0.20 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr12_-_48671612 | 0.20 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr7_+_5905091 | 0.20 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr2_+_9946121 | 0.19 |
ENSDART00000100696
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr7_-_31794476 | 0.19 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr2_-_51059691 | 0.19 |
ENSDART00000158131
|
ftr87
|
finTRIM family, member 87 |
| chr5_+_36439405 | 0.19 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr15_+_26940569 | 0.18 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr11_-_18604542 | 0.18 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_-_3328097 | 0.18 |
ENSDART00000193140
|
tmem184bb
|
transmembrane protein 184bb |
| chr11_-_18604317 | 0.18 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr7_-_29115772 | 0.17 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr14_+_1687357 | 0.16 |
ENSDART00000174675
|
hrh2a
|
histamine receptor H2a |
| chr11_+_45436703 | 0.16 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr23_-_4930229 | 0.16 |
ENSDART00000189456
|
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr17_+_23556764 | 0.15 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr1_+_12394205 | 0.15 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr8_-_11146687 | 0.15 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr17_+_6382275 | 0.15 |
ENSDART00000056324
|
CR628323.2
|
|
| chr5_-_68074592 | 0.15 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr7_-_73845736 | 0.15 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr7_+_5910467 | 0.14 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr18_+_44795711 | 0.14 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr8_+_53269657 | 0.14 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr3_-_62403550 | 0.14 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr20_+_21391181 | 0.14 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr19_+_2619444 | 0.14 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_-_9644630 | 0.14 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr8_-_39884359 | 0.14 |
ENSDART00000131372
|
mlec
|
malectin |
| chr7_-_18617187 | 0.13 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr18_+_21273749 | 0.13 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr23_+_37185247 | 0.11 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr7_+_5976613 | 0.11 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr5_-_2721686 | 0.10 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr2_-_25120197 | 0.10 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr19_+_5640504 | 0.10 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr23_-_2037566 | 0.09 |
ENSDART00000191312
ENSDART00000127443 |
prdm5
|
PR domain containing 5 |
| chr1_-_54063520 | 0.09 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr6_+_2951645 | 0.09 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr11_+_440305 | 0.09 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr6_+_102506 | 0.08 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr7_+_17953589 | 0.08 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr14_-_30945515 | 0.08 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr3_-_3327573 | 0.08 |
ENSDART00000136319
ENSDART00000166667 |
tmem184bb
|
transmembrane protein 184bb |
| chr11_-_15297129 | 0.08 |
ENSDART00000167985
ENSDART00000163031 ENSDART00000165010 ENSDART00000171250 ENSDART00000010684 ENSDART00000191714 ENSDART00000191164 |
rpn2
|
ribophorin II |
| chr19_+_791538 | 0.07 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr15_-_19677511 | 0.07 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ybx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 1.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.6 | GO:0010935 | dendritic cell antigen processing and presentation(GO:0002468) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 0.6 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.6 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.6 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.4 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.8 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.3 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.5 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.2 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.2 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.3 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |