Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
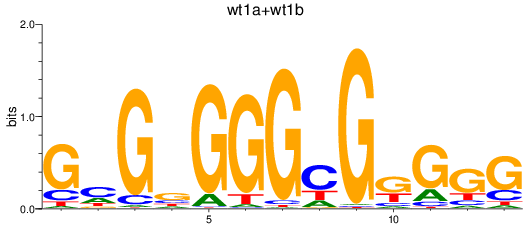
Results for wt1a+wt1b
Z-value: 2.02
Transcription factors associated with wt1a+wt1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
wt1b
|
ENSDARG00000007990 | WT1 transcription factor b |
|
wt1a
|
ENSDARG00000031420 | WT1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| wt1a | dr11_v1_chr25_-_15214161_15214161 | 0.71 | 6.7e-04 | Click! |
| wt1b | dr11_v1_chr18_-_45617146_45617146 | -0.07 | 7.8e-01 | Click! |
Activity profile of wt1a+wt1b motif
Sorted Z-values of wt1a+wt1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_43056153 | 6.75 |
ENSDART00000186377
|
prnpa
|
prion protein a |
| chr4_+_68562464 | 6.49 |
ENSDART00000192954
|
BX548011.4
|
|
| chr2_-_22535 | 6.10 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr20_-_25518488 | 5.99 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr7_-_10606 | 5.53 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr5_+_36614196 | 5.17 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr25_+_10811551 | 5.03 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr25_+_24250247 | 4.63 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr1_-_59422880 | 4.57 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr9_-_54001502 | 4.44 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr7_+_67325933 | 4.43 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr8_+_2656231 | 4.20 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr23_-_637347 | 4.13 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr4_-_68563862 | 4.11 |
ENSDART00000182970
|
BX548011.2
|
|
| chr17_+_27456804 | 4.01 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr11_+_6819050 | 3.90 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr2_+_58221163 | 3.80 |
ENSDART00000157939
|
FO704813.1
|
|
| chr8_-_22739757 | 3.76 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr9_-_33725972 | 3.74 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr21_-_39058490 | 3.66 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr23_-_3703569 | 3.65 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr15_-_37543591 | 3.62 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr8_+_26268100 | 3.62 |
ENSDART00000138882
|
slc26a6
|
solute carrier family 26, member 6 |
| chr3_-_11624694 | 3.61 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr21_+_1143141 | 3.50 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr13_-_638485 | 3.44 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr12_+_48744016 | 3.41 |
ENSDART00000175518
|
LO017648.1
|
|
| chr13_-_27767330 | 3.41 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr21_+_11468642 | 3.38 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr3_+_32526799 | 3.37 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr18_+_3004572 | 3.32 |
ENSDART00000190328
|
CABZ01002690.1
|
|
| chr7_-_6592142 | 3.30 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr25_+_34984333 | 3.28 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr3_+_19299309 | 3.28 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr11_+_36989696 | 3.25 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr19_+_41701660 | 3.22 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr5_+_6617401 | 3.22 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr5_+_9348284 | 3.21 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr23_+_44665230 | 3.21 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr4_-_77557279 | 3.19 |
ENSDART00000180113
|
AL935186.10
|
|
| chr7_+_10701938 | 3.16 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr16_-_22022456 | 3.13 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr18_+_660578 | 3.11 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr6_-_52428826 | 2.99 |
ENSDART00000047399
|
mmp24
|
matrix metallopeptidase 24 |
| chr9_-_30363770 | 2.97 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr7_-_57949117 | 2.96 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr8_+_8166285 | 2.94 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr9_-_54840124 | 2.92 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr11_-_103136 | 2.89 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr6_+_2271559 | 2.86 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr2_+_48073972 | 2.80 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr4_+_55758103 | 2.78 |
ENSDART00000185964
|
CT583728.23
|
|
| chr23_-_30785382 | 2.63 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr16_-_280835 | 2.63 |
ENSDART00000190541
|
LO017917.1
|
|
| chr14_-_2933185 | 2.62 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr6_-_60104628 | 2.61 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr14_+_30910114 | 2.60 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr8_+_26268726 | 2.55 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr11_-_44030962 | 2.55 |
ENSDART00000171910
|
FP016005.1
|
|
| chr5_-_13086616 | 2.53 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr13_+_13693722 | 2.52 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr1_+_54677173 | 2.49 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr25_+_35774544 | 2.48 |
ENSDART00000034737
ENSDART00000188162 |
cpne8
|
copine VIII |
| chr3_+_50312422 | 2.48 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr16_+_36650984 | 2.45 |
ENSDART00000156780
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr22_-_18112374 | 2.45 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr5_+_51443009 | 2.45 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr13_-_31166544 | 2.44 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr17_+_44756247 | 2.43 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr13_-_49666615 | 2.42 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr24_-_38110779 | 2.41 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr10_+_44940693 | 2.41 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr22_-_600016 | 2.40 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr25_-_207214 | 2.40 |
ENSDART00000193448
|
FP236318.3
|
|
| chr6_+_58492201 | 2.36 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr19_+_164013 | 2.32 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr6_+_6491013 | 2.32 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr5_+_9382301 | 2.32 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr3_-_30861177 | 2.31 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr17_-_20711735 | 2.29 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr10_+_32561317 | 2.27 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr15_-_33925851 | 2.26 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr20_+_46741074 | 2.25 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr21_+_11468934 | 2.24 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr3_-_62380146 | 2.24 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr19_+_935565 | 2.22 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr25_+_22683640 | 2.17 |
ENSDART00000131993
|
ush1c
|
Usher syndrome 1C |
| chr17_-_22048233 | 2.17 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr11_-_32723851 | 2.17 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr14_-_2369849 | 2.15 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr16_-_13613475 | 2.12 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr10_-_24343507 | 2.12 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_-_68022631 | 2.11 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr2_+_7192966 | 2.09 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr11_+_40649412 | 2.09 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr10_+_8885769 | 2.09 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr3_-_1190132 | 2.08 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr13_+_1100197 | 2.06 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr21_+_28958471 | 2.06 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_-_54639964 | 2.06 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr12_+_2522642 | 2.04 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr21_-_22485847 | 2.04 |
ENSDART00000162860
|
myo5b
|
myosin VB |
| chr23_+_29908348 | 2.04 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr25_-_19395476 | 2.01 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_+_48111285 | 2.01 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr2_-_57076687 | 1.99 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr19_+_33701366 | 1.99 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_-_18446483 | 1.98 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr25_+_35706493 | 1.96 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr7_+_74141297 | 1.96 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr20_-_1268863 | 1.95 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr5_+_37343643 | 1.94 |
ENSDART00000163993
ENSDART00000164459 ENSDART00000167006 ENSDART00000128122 ENSDART00000183720 |
klhl13
|
kelch-like family member 13 |
| chr3_-_49504023 | 1.92 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr11_+_25459697 | 1.91 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr11_+_41560792 | 1.91 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr21_+_11139175 | 1.90 |
ENSDART00000169024
|
agxt2
|
alanine--glyoxylate aminotransferase 2 |
| chr19_+_33701734 | 1.89 |
ENSDART00000123270
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_+_6576940 | 1.88 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr14_-_1565317 | 1.88 |
ENSDART00000169496
|
CABZ01064248.1
|
|
| chr23_+_43834376 | 1.86 |
ENSDART00000148989
|
si:ch211-149b19.4
|
si:ch211-149b19.4 |
| chr16_+_1803462 | 1.85 |
ENSDART00000183974
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr5_-_31928913 | 1.84 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr2_+_33382648 | 1.83 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr22_+_883678 | 1.82 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr15_-_30816370 | 1.81 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr19_+_233143 | 1.80 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr4_-_38476901 | 1.78 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr5_-_46273938 | 1.78 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr8_+_22485956 | 1.76 |
ENSDART00000099960
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr15_-_42736433 | 1.75 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr8_+_36666789 | 1.74 |
ENSDART00000144255
|
magixb
|
MAGI family member, X-linked b |
| chr1_-_37087966 | 1.73 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr19_+_63567 | 1.73 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr5_-_6433577 | 1.72 |
ENSDART00000160439
|
CABZ01037298.1
|
|
| chr3_+_7617353 | 1.72 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr18_-_33344 | 1.72 |
ENSDART00000129125
|
pde8a
|
phosphodiesterase 8A |
| chr18_-_1258777 | 1.72 |
ENSDART00000077106
ENSDART00000129065 |
ugt5f1
|
UDP glucuronosyltransferase 5 family, polypeptide F1 |
| chr10_+_10738880 | 1.72 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr19_-_6193448 | 1.71 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr14_-_12837432 | 1.70 |
ENSDART00000178444
|
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr20_-_52928541 | 1.69 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_-_411331 | 1.69 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr3_+_32526263 | 1.68 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_+_49232559 | 1.68 |
ENSDART00000154285
ENSDART00000175147 |
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr22_+_1539373 | 1.67 |
ENSDART00000172104
|
si:ch211-255f4.7
|
si:ch211-255f4.7 |
| chr12_-_48943467 | 1.66 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr21_-_24865454 | 1.65 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr25_-_37501371 | 1.64 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr9_+_48943471 | 1.64 |
ENSDART00000125090
|
stk39
|
serine threonine kinase 39 |
| chr21_-_22114625 | 1.62 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr16_+_4838808 | 1.61 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr10_+_439692 | 1.58 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr9_-_2572790 | 1.55 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr16_-_42152145 | 1.54 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr20_+_715739 | 1.54 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr10_+_45128375 | 1.53 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr2_-_9646857 | 1.53 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr23_+_27912079 | 1.52 |
ENSDART00000171859
|
BX539336.1
|
|
| chr14_-_12837052 | 1.52 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr17_+_46387086 | 1.52 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr14_+_22172047 | 1.51 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_+_23165749 | 1.50 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr7_-_29534001 | 1.50 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr5_-_67145505 | 1.50 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr15_+_47418565 | 1.48 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr7_-_51627019 | 1.47 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr3_+_22079219 | 1.46 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr8_+_51958968 | 1.45 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr19_-_617246 | 1.44 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr16_+_10918252 | 1.44 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr6_-_58828113 | 1.44 |
ENSDART00000180934
|
kif5ab
|
kinesin family member 5A, b |
| chr3_-_36944749 | 1.43 |
ENSDART00000154501
|
cntnap1
|
contactin associated protein 1 |
| chr2_+_48074243 | 1.42 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr8_-_53490376 | 1.41 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr24_-_38644937 | 1.40 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr11_-_97817 | 1.39 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr17_-_19828505 | 1.38 |
ENSDART00000154745
|
fmn2a
|
formin 2a |
| chr9_+_13714379 | 1.38 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr17_+_11507131 | 1.36 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr6_-_30859656 | 1.36 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr4_+_3980247 | 1.34 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_+_7740004 | 1.34 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_-_24068848 | 1.34 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr2_-_11912347 | 1.34 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr20_+_13969414 | 1.34 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr12_+_48511605 | 1.34 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr3_+_12322170 | 1.33 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr23_+_19564392 | 1.33 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr20_-_51727860 | 1.33 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr6_-_7776612 | 1.32 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_+_38924975 | 1.32 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr11_+_45461419 | 1.31 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr9_-_1434484 | 1.30 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr12_-_314899 | 1.30 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr10_-_33156789 | 1.30 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr6_-_442163 | 1.29 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr6_-_16406210 | 1.29 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr14_+_2243 | 1.29 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr14_-_962171 | 1.27 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr16_-_12953739 | 1.27 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
Network of associatons between targets according to the STRING database.
First level regulatory network of wt1a+wt1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.0 | 6.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 1.0 | 4.0 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.8 | 2.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.6 | 3.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.6 | 2.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.6 | 3.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 2.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.6 | 0.6 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 3.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.5 | 2.0 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 3.3 | GO:0036268 | swimming(GO:0036268) |
| 0.5 | 1.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.5 | 1.8 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.4 | 1.7 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.4 | 6.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 3.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 2.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.3 | 2.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.3 | 7.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 5.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 1.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.3 | 2.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 2.0 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 1.3 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 1.0 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 3.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 3.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 5.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.2 | 1.9 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 1.7 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.2 | 5.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 1.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 0.8 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 3.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 1.9 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.2 | 2.0 | GO:0003139 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.2 | 0.5 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 4.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 0.5 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 2.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 0.5 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.2 | 1.2 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 2.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.4 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 1.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 2.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 2.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 3.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 2.0 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.0 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 1.3 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 2.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.8 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 2.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.0 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.8 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 2.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 3.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 4.9 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.6 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.4 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 3.2 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.2 | GO:0031936 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 0.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 1.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 3.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 2.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.5 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 1.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 5.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 2.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 1.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 1.4 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 2.4 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 2.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.1 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.7 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 1.1 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.7 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 5.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 2.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.1 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.4 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 1.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.1 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.9 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 2.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 5.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 2.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 2.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.2 | 4.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 2.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 3.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 1.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 5.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.2 | 2.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 2.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 5.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 6.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 4.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 4.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 3.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 6.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.9 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 5.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.8 | 3.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.7 | 3.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 2.0 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.6 | 4.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.6 | 2.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 1.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 3.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 2.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 1.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 1.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 1.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 1.1 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 3.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 2.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 2.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 2.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 6.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 6.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 2.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.3 | 2.1 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 3.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 1.0 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 0.8 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 4.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 0.5 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 3.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 8.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 4.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 1.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 2.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 3.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 3.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 4.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.7 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 3.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 3.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 3.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 5.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 2.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 3.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.8 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 6.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 7.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 5.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 6.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |