Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
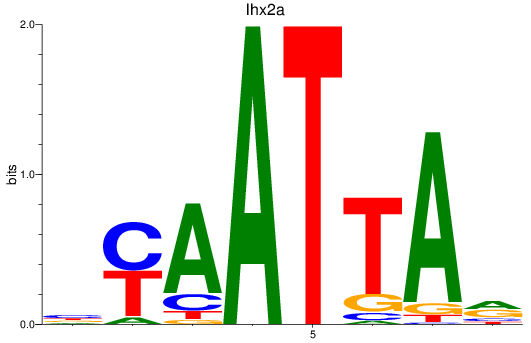
Results for vsx1_shox_shox2_uncx4.1_lhx2a
Z-value: 0.89
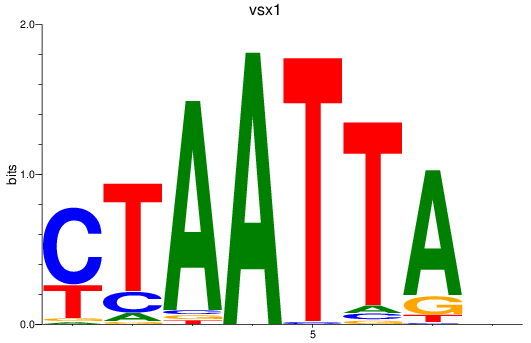
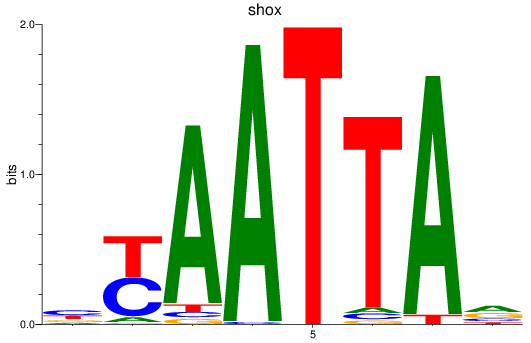
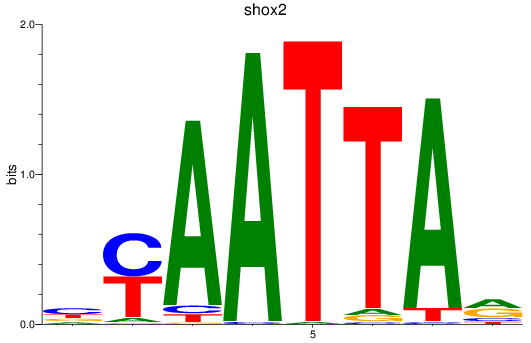
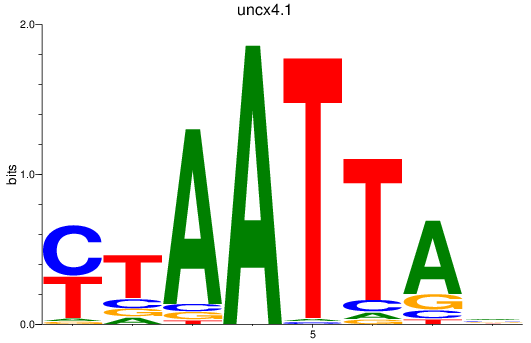
Transcription factors associated with vsx1_shox_shox2_uncx4.1_lhx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vsx1
|
ENSDARG00000056292 | visual system homeobox 1 homolog, chx10-like |
|
vsx1
|
ENSDARG00000109766 | visual system homeobox 1 homolog, chx10-like |
|
shox
|
ENSDARG00000025891 | short stature homeobox |
|
shox2
|
ENSDARG00000075713 | short stature homeobox 2 |
|
uncx4.1
|
ENSDARG00000037760 | Unc4.1 homeobox (C. elegans) |
|
lhx2a
|
ENSDARG00000037964 | LIM homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vsx1 | dr11_v1_chr17_-_21066075_21066105 | -0.51 | 2.6e-02 | Click! |
| uncx4.1 | dr11_v1_chr1_+_9290103_9290103 | 0.45 | 5.5e-02 | Click! |
| shox2 | dr11_v1_chr15_-_2188332_2188422 | -0.41 | 8.2e-02 | Click! |
| shox | dr11_v1_chr9_+_34641237_34641237 | -0.32 | 1.9e-01 | Click! |
| lhx2a | dr11_v1_chr21_-_8420830_8420830 | 0.19 | 4.3e-01 | Click! |
Activity profile of vsx1_shox_shox2_uncx4.1_lhx2a motif
Sorted Z-values of vsx1_shox_shox2_uncx4.1_lhx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_26596794 | 3.61 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr4_+_306036 | 2.67 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr22_+_18315490 | 2.49 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr2_+_20793982 | 2.40 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr13_+_22295905 | 2.00 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr5_-_12219572 | 1.96 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr12_-_35830625 | 1.94 |
ENSDART00000180028
|
CU459056.1
|
|
| chr19_+_15440841 | 1.91 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_30668580 | 1.80 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_-_20372977 | 1.72 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr23_-_17003533 | 1.71 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr17_-_16422654 | 1.61 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr18_-_33979693 | 1.60 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr2_-_55298075 | 1.55 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_-_4528326 | 1.53 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr19_+_15441022 | 1.53 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr16_-_16761164 | 1.52 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr16_+_29509133 | 1.50 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr24_+_19415124 | 1.49 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr2_+_41526904 | 1.49 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr25_+_14507567 | 1.46 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr6_-_43283122 | 1.46 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr9_+_25776971 | 1.40 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr22_+_16535575 | 1.40 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr15_+_34988148 | 1.36 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr5_+_60590796 | 1.35 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr8_-_50888806 | 1.30 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr15_+_17258246 | 1.25 |
ENSDART00000101707
|
dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr20_-_9095105 | 1.17 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr16_+_28994709 | 1.12 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr8_+_21353878 | 1.09 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr18_+_24921587 | 1.07 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr15_-_16177603 | 1.05 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr6_-_40922971 | 1.04 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr2_-_7246848 | 1.02 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr20_-_22476255 | 1.02 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_56954002 | 0.98 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr6_+_41191482 | 0.98 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr10_+_6884627 | 0.96 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr2_-_38363017 | 0.95 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr23_-_33709964 | 0.94 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr15_+_1796313 | 0.94 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr4_-_9891874 | 0.92 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr23_+_38251864 | 0.91 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr11_+_33818179 | 0.91 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr5_+_66433287 | 0.90 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr20_-_48485354 | 0.88 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr18_-_2433011 | 0.87 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr8_+_23355484 | 0.86 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr20_-_23426339 | 0.86 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_-_6078222 | 0.85 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_-_35171162 | 0.85 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr2_-_38206034 | 0.84 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr13_+_27232848 | 0.84 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr3_-_16719244 | 0.83 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr17_-_31659670 | 0.83 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr25_-_20378721 | 0.82 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr21_-_37973819 | 0.82 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr11_+_18873113 | 0.80 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr18_-_15551360 | 0.80 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr1_+_10318089 | 0.80 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr6_+_39232245 | 0.79 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr16_+_25116827 | 0.79 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr23_+_39695827 | 0.79 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr13_-_31017960 | 0.78 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr18_-_25568994 | 0.78 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr5_-_30481263 | 0.78 |
ENSDART00000086734
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr6_-_8311044 | 0.78 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr14_-_22113600 | 0.77 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr23_-_31913231 | 0.76 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr24_-_25144441 | 0.76 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr20_-_28642061 | 0.76 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr1_-_5455498 | 0.76 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr13_-_31008275 | 0.76 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr17_-_49438873 | 0.75 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr14_+_45406299 | 0.75 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr6_-_43616936 | 0.74 |
ENSDART00000149301
|
foxp1b
|
forkhead box P1b |
| chr3_-_26183699 | 0.74 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr19_+_2631565 | 0.74 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_+_21731382 | 0.73 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr3_-_21094437 | 0.73 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr1_-_55248496 | 0.73 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr18_+_17827149 | 0.70 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr17_+_21295132 | 0.69 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr1_+_34696503 | 0.69 |
ENSDART00000186106
|
CR339054.2
|
|
| chr8_-_23612462 | 0.69 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr3_-_23643751 | 0.68 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr9_-_35633827 | 0.68 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr17_+_16046314 | 0.67 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_+_16584382 | 0.66 |
ENSDART00000112039
|
CR790388.1
|
|
| chr23_-_18130264 | 0.66 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr19_+_22062202 | 0.66 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr8_+_11425048 | 0.65 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr5_-_25733745 | 0.65 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr15_-_2493771 | 0.65 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr19_-_19871211 | 0.65 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr23_+_11285662 | 0.65 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr13_+_30903816 | 0.64 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr21_-_40676224 | 0.64 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr13_+_38817871 | 0.64 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr5_+_68807170 | 0.64 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr6_-_12172424 | 0.63 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr4_+_31405646 | 0.63 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr23_-_31913069 | 0.63 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr2_+_33326522 | 0.63 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr7_+_24023653 | 0.63 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr21_+_25802190 | 0.62 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr21_+_34088110 | 0.62 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr18_+_19456648 | 0.62 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr24_-_40860603 | 0.62 |
ENSDART00000188032
|
CU633479.7
|
|
| chr21_-_18648861 | 0.61 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr4_+_13586689 | 0.61 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr12_-_43664682 | 0.60 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr24_+_9412450 | 0.60 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr5_-_14326959 | 0.60 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr19_-_30510259 | 0.60 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr8_+_17168114 | 0.59 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr6_+_40922572 | 0.59 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_30488063 | 0.58 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr10_-_21362320 | 0.58 |
ENSDART00000189789
|
avd
|
avidin |
| chr9_-_32753535 | 0.58 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr2_+_4146606 | 0.57 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr3_-_46811611 | 0.57 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr24_-_34680956 | 0.57 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr11_-_29563437 | 0.57 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr19_-_19379084 | 0.57 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr25_+_35891342 | 0.56 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr2_+_6253246 | 0.56 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_-_2450597 | 0.56 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr17_+_21964472 | 0.56 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr17_+_24821627 | 0.55 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr17_+_12865746 | 0.55 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr12_-_33972798 | 0.55 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr2_-_38284648 | 0.55 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr10_-_21362071 | 0.55 |
ENSDART00000125167
|
avd
|
avidin |
| chr16_+_54209504 | 0.55 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr20_+_539852 | 0.55 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr5_-_41307550 | 0.55 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr19_+_19786117 | 0.54 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr3_+_18807006 | 0.53 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr15_+_27384798 | 0.52 |
ENSDART00000164887
|
tbx4
|
T-box 4 |
| chr2_+_4146299 | 0.52 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr4_+_11723852 | 0.52 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr4_+_9400012 | 0.51 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr10_+_17714866 | 0.51 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr23_-_29553430 | 0.51 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr24_+_1023839 | 0.51 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr5_+_38900249 | 0.51 |
ENSDART00000097856
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr3_+_32365811 | 0.50 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr17_-_41798856 | 0.50 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr7_+_29167744 | 0.49 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr10_+_43039947 | 0.49 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr6_-_8392104 | 0.48 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr7_+_25221757 | 0.48 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr3_+_26244353 | 0.48 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr23_-_29553691 | 0.48 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr10_+_17371356 | 0.47 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr15_-_20024205 | 0.47 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr17_-_2690083 | 0.47 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr1_-_36152131 | 0.47 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr7_-_31759602 | 0.47 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr17_+_23298928 | 0.47 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr1_+_30100257 | 0.47 |
ENSDART00000134311
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr10_-_33297864 | 0.47 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr7_-_31759394 | 0.46 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr21_+_30746348 | 0.45 |
ENSDART00000050172
|
trpc2b
|
transient receptor potential cation channel, subfamily C, member 2b |
| chr14_-_33945692 | 0.45 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr3_-_58798815 | 0.45 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr10_+_17235370 | 0.45 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr17_-_10122204 | 0.44 |
ENSDART00000160751
|
BX088587.1
|
|
| chr9_+_24008879 | 0.44 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr7_-_30174882 | 0.44 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr21_+_15592426 | 0.44 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr2_-_28085884 | 0.44 |
ENSDART00000131506
|
cdh6
|
cadherin 6 |
| chr11_-_36341028 | 0.43 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr17_+_1496107 | 0.43 |
ENSDART00000187804
|
LO018430.1
|
|
| chr3_-_26787430 | 0.43 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr14_+_35428152 | 0.43 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr9_+_7732714 | 0.43 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr21_-_37194365 | 0.42 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr1_-_40102836 | 0.42 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr9_+_38457806 | 0.42 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr9_+_38372216 | 0.42 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr9_+_25568839 | 0.41 |
ENSDART00000177342
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr8_-_37249813 | 0.41 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr14_+_22113331 | 0.41 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr8_-_15129573 | 0.41 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr5_-_68333081 | 0.41 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr14_+_34971554 | 0.41 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr22_+_5176255 | 0.41 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr22_-_8725768 | 0.40 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr11_+_24820542 | 0.40 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr8_-_15292197 | 0.40 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr19_+_21362553 | 0.40 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr17_+_16046132 | 0.40 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr21_-_25801956 | 0.40 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr13_+_22476742 | 0.40 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr5_+_6954162 | 0.39 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr23_-_1017605 | 0.39 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr10_+_16092671 | 0.39 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx1_shox_shox2_uncx4.1_lhx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.7 | 2.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.5 | 1.4 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 2.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 1.6 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.4 | 1.5 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.4 | 1.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 1.7 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 1.8 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 1.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 0.9 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 1.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.0 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 2.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.7 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.6 | GO:0048890 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.1 | 1.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.6 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.4 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.3 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.1 | 0.7 | GO:0060114 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 1.3 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.6 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.2 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.3 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.0 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.0 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.3 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.1 | 1.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.5 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 1.0 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.1 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.2 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 1.5 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.5 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0032042 | mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.0 | 0.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.2 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.5 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 1.4 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.0 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.5 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) negative regulation of nervous system development(GO:0051961) |
| 0.0 | 0.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.6 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 1.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.9 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 0.9 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.6 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 2.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 1.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.4 | 1.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 0.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 2.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 1.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 2.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 0.5 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 1.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 3.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H4-K20 specific)(GO:0035575) histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |