Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
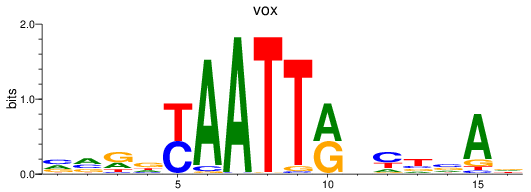
Results for vox
Z-value: 0.49
Transcription factors associated with vox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vox
|
ENSDARG00000099761 | ventral homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vox | dr11_v1_chr13_-_50624743_50624743 | 0.56 | 1.3e-02 | Click! |
Activity profile of vox motif
Sorted Z-values of vox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_48667056 | 1.19 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr8_-_39822917 | 0.89 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr3_-_15444396 | 0.84 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr11_-_33868881 | 0.83 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr10_+_32104305 | 0.76 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr16_+_54209504 | 0.73 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr10_-_35257458 | 0.71 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr11_-_6877973 | 0.71 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr10_-_8046764 | 0.67 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr22_-_36530902 | 0.59 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr16_-_7443388 | 0.58 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr17_+_16046314 | 0.57 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_-_15999501 | 0.57 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_+_61459422 | 0.53 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr4_+_35553514 | 0.53 |
ENSDART00000182938
|
CR847906.1
|
|
| chr8_-_50888806 | 0.52 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr23_+_35713557 | 0.51 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr4_+_5506952 | 0.50 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr24_+_19415124 | 0.48 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr21_+_22892836 | 0.46 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr3_+_45365098 | 0.46 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr24_+_36204028 | 0.45 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr17_+_17764979 | 0.45 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr20_+_26966725 | 0.45 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr21_-_15200556 | 0.45 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr3_-_23643751 | 0.44 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr6_-_7720332 | 0.43 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr19_-_10323845 | 0.43 |
ENSDART00000151259
ENSDART00000151821 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr2_-_22363460 | 0.43 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr9_+_21358941 | 0.42 |
ENSDART00000147619
ENSDART00000059402 |
eef1akmt1
|
EEF1A lysine methyltransferase 1 |
| chr18_+_35173683 | 0.42 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr19_-_10971230 | 0.42 |
ENSDART00000166196
|
LO018584.1
|
|
| chr12_-_18414536 | 0.41 |
ENSDART00000132956
ENSDART00000078724 |
atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr13_-_33227411 | 0.41 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr14_+_30568961 | 0.40 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr10_+_7593185 | 0.40 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr15_+_15856178 | 0.39 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr6_+_15762647 | 0.38 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr21_+_31253048 | 0.37 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr9_+_21402863 | 0.36 |
ENSDART00000125357
|
cx30.3
|
connexin 30.3 |
| chr16_+_42471455 | 0.35 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr5_-_25733745 | 0.35 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr8_-_49764156 | 0.35 |
ENSDART00000177589
ENSDART00000189142 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr18_+_48423973 | 0.34 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr11_+_11303458 | 0.34 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr6_-_52348562 | 0.33 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr22_+_2198246 | 0.33 |
ENSDART00000165168
ENSDART00000157485 ENSDART00000168713 |
znf1157
znf1166
|
zinc finger protein 1157 zinc finger protein 1166 |
| chr11_-_16152400 | 0.33 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr4_+_14727212 | 0.32 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr3_+_45364849 | 0.32 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr22_+_16308450 | 0.31 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr6_-_39275793 | 0.31 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr1_+_22691256 | 0.31 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr10_+_28160265 | 0.30 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr6_-_40922971 | 0.30 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr8_+_17078692 | 0.30 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr23_+_45966436 | 0.30 |
ENSDART00000172160
|
CABZ01069338.1
|
|
| chr5_+_66353750 | 0.29 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr5_-_51484156 | 0.29 |
ENSDART00000162064
|
CR388055.1
|
|
| chr7_-_34262080 | 0.29 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr11_+_16152316 | 0.29 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr5_+_66433287 | 0.29 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr12_+_22580579 | 0.28 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr23_+_32021803 | 0.28 |
ENSDART00000012963
|
trappc6b
|
trafficking protein particle complex 6b |
| chr10_+_35257651 | 0.27 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr2_-_10896745 | 0.27 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr3_-_16719244 | 0.26 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr4_-_49119557 | 0.26 |
ENSDART00000150263
|
znf1036
|
zinc finger protein 1036 |
| chr10_+_33744098 | 0.26 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr8_+_11325310 | 0.25 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr5_+_29671681 | 0.25 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr15_+_23528010 | 0.25 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr9_+_43799829 | 0.24 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr24_+_6107901 | 0.24 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr14_+_11290828 | 0.24 |
ENSDART00000184078
|
rlim
|
ring finger protein, LIM domain interacting |
| chr2_-_23411368 | 0.22 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr4_+_57580303 | 0.22 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr21_-_17296789 | 0.22 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr10_-_35149513 | 0.22 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr13_+_8677166 | 0.22 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr9_-_12594759 | 0.21 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr11_-_7380674 | 0.20 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr19_+_26718074 | 0.20 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr16_-_17197546 | 0.19 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr4_+_49322310 | 0.19 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr18_+_32444636 | 0.19 |
ENSDART00000164223
|
v2ra1
|
vomeronasal 2 receptor, a1 |
| chr16_+_20056030 | 0.19 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr10_+_39091353 | 0.19 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr5_-_29671586 | 0.18 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr5_+_66353589 | 0.18 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr21_+_20396858 | 0.18 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr24_+_38301080 | 0.18 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr3_-_53722182 | 0.18 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr3_-_36690348 | 0.18 |
ENSDART00000192513
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr4_-_41712014 | 0.18 |
ENSDART00000138165
|
znf976
|
zinc finger protein 976 |
| chr16_+_19033554 | 0.17 |
ENSDART00000182430
|
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr20_-_37813863 | 0.17 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_-_42878178 | 0.17 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr6_+_19383267 | 0.17 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr3_-_38783951 | 0.17 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr21_-_32097908 | 0.17 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr20_-_49704915 | 0.17 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr19_+_31541607 | 0.16 |
ENSDART00000181604
|
gmnn
|
geminin, DNA replication inhibitor |
| chr4_+_14727018 | 0.16 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr11_+_31864921 | 0.16 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr4_-_41269844 | 0.16 |
ENSDART00000186177
|
CR388165.2
|
|
| chr25_+_31868268 | 0.16 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr4_-_55459037 | 0.16 |
ENSDART00000172184
|
znf569l
|
zinc finger protein 569, like |
| chr8_+_11471350 | 0.16 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr20_-_34663209 | 0.15 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr6_-_54815886 | 0.15 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr11_+_2391649 | 0.15 |
ENSDART00000104571
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr15_+_20344670 | 0.15 |
ENSDART00000158986
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_-_7567815 | 0.14 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr2_+_30787128 | 0.14 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr25_-_18953322 | 0.14 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr10_+_11265387 | 0.14 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr16_+_20055878 | 0.14 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr18_-_40708537 | 0.14 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr6_+_32382743 | 0.14 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr25_+_5035343 | 0.14 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr1_-_55263736 | 0.14 |
ENSDART00000152504
ENSDART00000152687 |
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr3_-_15210491 | 0.14 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr22_-_17474781 | 0.13 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr21_+_40195452 | 0.13 |
ENSDART00000108650
|
or115-15
|
odorant receptor, family F, subfamily 115, member 15 |
| chr19_+_10592778 | 0.13 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr22_-_17474583 | 0.12 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr11_+_24703108 | 0.12 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr1_-_11596829 | 0.12 |
ENSDART00000140725
|
si:dkey-26i13.7
|
si:dkey-26i13.7 |
| chr3_+_22334012 | 0.12 |
ENSDART00000193526
|
ifnphi3
|
interferon phi 3 |
| chr2_+_33747509 | 0.12 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr10_-_28028998 | 0.12 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr5_+_56268436 | 0.12 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr15_-_590787 | 0.11 |
ENSDART00000189367
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr8_-_25874426 | 0.11 |
ENSDART00000142730
|
opn7c
|
opsin 7, group member c |
| chr23_-_39772067 | 0.11 |
ENSDART00000159519
ENSDART00000189046 |
im:6912630
|
im:6912630 |
| chr15_-_18200358 | 0.10 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr8_-_22274222 | 0.10 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr7_-_59210882 | 0.09 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr2_+_30786773 | 0.09 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr12_-_19119176 | 0.09 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr1_+_27024068 | 0.08 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr7_-_45999333 | 0.08 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr11_-_29737088 | 0.07 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr22_-_7457247 | 0.07 |
ENSDART00000106081
|
BX511034.2
|
|
| chr15_+_5360407 | 0.07 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr5_+_7989210 | 0.07 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr16_-_39477509 | 0.06 |
ENSDART00000191330
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr25_-_21507676 | 0.06 |
ENSDART00000010706
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr17_+_41992054 | 0.06 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr21_+_6751760 | 0.06 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr2_-_15324837 | 0.06 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr2_+_22363824 | 0.05 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr7_-_22699716 | 0.05 |
ENSDART00000193773
|
BX470211.5
|
|
| chr21_-_30032706 | 0.05 |
ENSDART00000156721
|
pwwp2a
|
PWWP domain containing 2A |
| chr23_-_9991060 | 0.04 |
ENSDART00000111518
|
plxnb1a
|
plexin b1a |
| chr4_-_50930346 | 0.04 |
ENSDART00000184245
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr20_+_45853154 | 0.04 |
ENSDART00000181109
|
AL929237.4
|
|
| chr21_+_15713097 | 0.04 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr7_-_40110140 | 0.04 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr15_-_2493771 | 0.04 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr15_-_15357178 | 0.03 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr1_+_27023687 | 0.03 |
ENSDART00000189093
|
bnc2
|
basonuclin 2 |
| chr21_+_21812311 | 0.03 |
ENSDART00000151253
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr19_-_7690975 | 0.02 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr17_+_23298928 | 0.02 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr1_+_57311901 | 0.02 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr1_-_7582859 | 0.02 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_-_49250490 | 0.02 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr15_+_41027466 | 0.01 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr13_+_33688474 | 0.00 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr17_-_45525936 | 0.00 |
ENSDART00000084102
|
hhipl2
|
HHIP-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.5 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.9 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.5 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0061217 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.5 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |